graphical stability analysis How to experimentally verify these ideas
advertisement

Review L6: Multistability & introduction λ phage model
λ phage model (Hasty et al.) as example for applying
mass action law.
K
1→ X
2X ←⎯
⎯
2
K1
λ λ
K2
OR3
λ λ
OR2
OR3
λ λ
λ λ
OR3
OR2
λ
K
2→ DX
⎯
D + X ←⎯
2
2
K
2→ DX
⎯
D + X ←⎯
2
2
λ
λ λ
OR2
K
1→ X
2X ←⎯
⎯
2
most important
K
step in
3 → DX*
D + X ←⎯
⎯
2
2
modeling !!
fast
K
4 → DX DX
DX + X ←⎯
⎯
2 2
2 2
k
t
DX + P ⎯⎯
⎯→ DX + P + nX
2
2
k
most important
d
X ⎯⎯
⎯→ A
slow
step in
modeling !!
biology
math
1
K
3 → DX*
D + X ←⎯
⎯
2
2
K
4 → DX DX
DX + X ←⎯
⎯
2 2
2 2
k
t → DX + P + nX
DX + P ⎯⎯
⎯
2
2
k
d→A
X ⎯⎯
⎯
mass action
dx
αx2
=
− γx + 1
dt 1 +(1 + σ )x2 + σ x4
1
2
K
σ = 3
1 K
relative binding
2
constants
K
σ = 4
2 K
2
nk p d
α = t 0 T ~ synthesis/basal
rate
r
k
d
γ=
~ degradation/basal
r K K
rate
1 2
choose elegant (dimensionless, relative) variables2 !
graphical stability analysis
How to experimentally verify these ideas ?
S th ti Biology
Synthetic
Bi l
Build your own designed network ‘from scratch’
and test your model
Isaacs et al. Prediction and measurement of an autoregulatory
genetic module.
module PNAS 100,
100 7714 (2003)
3
4
IIsaacs et al.
l Prediction
P di i andd measurement off an autoregulatory
l
genetic module. PNAS 100, 7714 (2003)
5
6
7
8
Example of cellular memory:
embryonic
b
i development
d
l
t
Transient stimuli produce persistent responses
How is this memory stored?
9
10
A simple mechanism for persistence: positive feedback
A dramatic example of
cellular
ce
u a dec
decision-making:
so
a g
stem cells
x
promoter
y
x
gene
promoter
x
y
x
y
+
y
x
y
y
t
Reya et al. Nature 414, 105 (2001)
gene
y
x
x
11
x
y
t
12
g(y)
g(y)
Y
f(y)
Y
Y
PCONST
steady
state
gene Y
0
dy
= f ( y) − g ( y)
dt
0
+
Y
gene Y
PY
f(y)
unstable
fixed
point
stable
fixed
point
i t
0
y
dy
= f ( y) − g ( y)
dt
f ( y ) = const.
g ( y ) = γy
f ( y) = α
y
0
y
Kd + y
g ( y ) = γy
13
g(y)
PY
The Expression Potential U: a Useful Concept from Classical Mechanics
Classical Mechanics
Y Y
Y Y
14
+
Y
unstable f(y)
fixed
point
stable
fixed
point
dx
dt
stable
fi d
fixed
point
0
gene Y
0
dy
= f ( y) − g ( y)
dt
0
m
inertia friction externall
forces
y2
f ( y) = α 2
Kd + y2
( )
U(x)
y: protein concentration
x
d 2x
d
dx
dU
m 2 +γ
= Fext ≡ −
dt
dx
dt
y
Chemical Kinetics
dx
1 dU
=−
dt
γ dx
dy
= f ( y) − g ( y)
dt
U ( y) = −∫
y
0
{ f ( y' ) − g ( y ' )}dy
d '
U(y)
f(y)-g(y)
Fext
g ( y ) = γy
15
x
y
16
g(y)
g(y)
Y
f(y)
Y
Y
PCONST
steady
state
gene Y
PY
Y
gene Y
0
o
[ f ( yy' ) − g ( y' )]dy '
stable
steady
state
t t
0
dy
= f ( y) − g ( y)
dt
f ( y) = α
y
Kd + y
g ( y ) = γy
potential U
f ( y ) = const.
g ( y ) = γy
U ( y) ≡ −∫
y
potential U
dy
= f ( y) − g ( y)
dt
f(y)
unstable
steady
state
+
U ( y ) ≡ − ∫ [ f ( y ' ) − g ( y ' )]dy '
y
o
y
17
g(y)
Y Y
Y Y
PY
+
Y
unstable f(y)
fixed
point
stable
fixed
point
y
18
A classic genetic switch: the lac operon
transcription is blocked by lac repressor (LacI)
stable
fi d
fixed
point
extracellular
intracellular
gene Y
RNA polymerase
0
f ( y) = α
g ( y ) = γy
y2
K d2 + y 2
lac repressor (LacI)
potential U
dy
= f ( y) − g ( y)
dt
Plac
lacZ
lacY
lacA
U ( y ) ≡ − ∫ [ f ( y ' ) − g ( y ' )]dy '
y
o
y
19
20
A classic genetic switch: the lac operon
A classic genetic switch: the lac operon
once LacI unbinds, RNA polymerase starts transcription
RNA polymerase transcribes lac genes
extracellular
intracellular
extracellular
intracellular
Permease
(LacY)
β-gal (LacZ)
lac repressor (LacI)
lac repressor (LacI)
RNA polymerase
Plac
lacZ
RNA polymerase
lacY
lacA
Plac
lacZ
lacY
lacA
21
A classic genetic switch: the lac operon
22
A classic genetic switch: the lac operon
intracellular lactose binds LacI resulting in inactive repressor
increased concentration of LacY results in synthesis of more LacY
extracellular
t
ll l llactose
t
extracellular
intracellular
extracellular
intracellular
intracellular lactose
Plac
lacZ
lacY
Plac
lacA
23
lacZ
lacY
lacA
24
Regulation of lactose uptake: a positive feedback system
Regulation of lactose uptake: a positive feedback system
extracellular
lactose
extracellular
lactose
glucose
P
Permease
(LacY)
intracellular
lactose
P
Permease
(LacY)
CRP
repressor
(LacI)
Plac
lacZ
lacY
lacA
repressor
(LacI)
Plac
lacZ
lacY
lacA
25
Positive feedback in a bacterial
regulatory network
x
LacY
R
dy
1
=α
−y
dt
1 + R / R0
τx
dx
= βy − x
dt
y
dy
= f −g
dt
0.4
dy/dt
τy
TMG
TMG
The lac system is bistable
R
1
=
RT 1 + ( x / x0 ) n
dy
1 + ( βy ) n
=α
−y
d
dt
ρ + ( βy ) n
0.2
0.0
n=1
-0.2
0.0
dy
= f ( y) − g ( y)
dt
0.5
1.0
1.5
2.0
y
LacI
y
Plac
lacZ
Plac
gfp
lacY lacA
GFP
φ ( y ) = − ∫ ( f − g )dy '
dy
1 + ( βy ) n
=α
−y
ρ + ( βy ) n
dt
0
φ (y)
β
26
maximal activation: α
0.0
extracellular TMG: β
0.5
1.0
1.5
2.0
y
repression factor: ρ =(1+RT/R0)-1
27
28
Network response can be either
discontinuous or continuous
The lac system is bistable
dy
= f −g
dt
dy/dt
0.2
dy
1 + ( βy ) n
−y
=α
d
dt
ρ + ( βy ) n
0.1
Continuous
Discontinuous
0.0
n=2
-0.1
0.0
0.5
1.0
1.5
2.0
Decrease
repression
factor
Phase diagram
y
φ ( y ) = − ∫ ( f − g )dy '
0.6
αβ / ρ
φ (y)
0
αβ/ρ
TMG ~ β
T
TMG ~ β
y
0.4
0.2
0.5
1.0
1.5
2.0
y
0.0
0 05 0.10
0.05
0 10 0.15
0 15 0.20
0 20
1/ρ
29
30
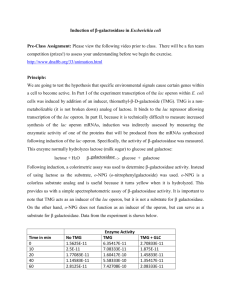
Experimental protocol:
Bistability allows memory storage
Measure single cell fluorescence histograms (both GFP and HcRed) in ‘steady-state’ as a
function of:
(i) external TMG concentration (continuous variable)
(ii) external glucose concentration (continuous variable)
(iii) initial condition (binary variable: fully induced versus not induced)
decrease TMG
Plac-GFP
G
is integrated in the chromosome; Pgat-HcRed is on a low copy plasmid
Pe
ermease
conce
entration (y)
0.0
Glu
TMG
cAMP
TMG
CRP
LacI
LacY
lactose
metabolism
LacZ
Extracellular TMG
Plac
lacZ
lacY lacA
Plac
gfp
Pgat
HcRed
GFP
HcRed
increase TMG
31
32
Induction protocol, history dependent experiments
> 24 hours
liquid culture
(~ 24 generations)
100
10
0 μM TMG
...
Green ffluoresccence
G
1 μM TMG
2 μM TMG
> 12 hours liquid culture
0 mM TMG
(all cells OFF)
split single
colony
1 mM TMG
0 μM TMG
1
100
initial LOW state
10
1
...
1 μM TMG
2 μM TMG
> 12 hours liquid culture
G
1 mM TMG
(all cells ON)
initial HIGH state
1 mM TMG
2
33
Novick and Weiner, PNAS 43, 553 (1957); Cohn and Horibata, J. Bacteriol. 78, 601 (1959)
4
6
8 10
Extracellular TMG (μM)
20
40
34
Mapping the bistable region as a function of TMG and glucose concentration
100
initial HIGH state
Green ffluoresccence
G
10
1
100
monostable
bistable
monostable
t bl
initial LOW state
10
1
2
4
6
8 10
Extracellular TMG (μM)
20
40
35
36
Functions α, β, and ρ are calculated from switching thresholds
Inducer exclusion:
TMG import rate does not only depend on extracellular TMG but also
depends on glucose level: β(T,G) = βT(T)βG(G)
1
⎧R
⎪ R = 1 + ( x/x )n
o
⎪ T
1
⎪ dy
=α
−y
⎨τ y
1
+
dt
R/R
o
⎪
⎪ dx
⎪τ x dt = βy − x
⎩
ρ = 1+RT/Ro
α: maximum
i
llacY
Y synthesis
h i rate
obtained if R → 0
α / ρ : minimum lacY synthesis rate
obtained if R → RT
37
Inducerr exclusion
n (βG)
TMG uptake rate (β T)
0.10
0.08
0.06
0 04
0.04
0.02
120
100
80
60
40
20
0
0.00
0
200
400
600
800 1000
0
40
60
80 100 120
inducer exclusion: glucose binds directly to LacY inhibiting TMG uptake
38
Inducer exclusion:
Network response can be either
discontinuous or continuous
TMG import rate does not only depend on extracellular TMG but also
depends on glucose level: β(T,G) = βT(T)βG(G)
Continuous
Discontinuous
βT(T)
Decrease
repression
factor
TMG
TMG
0.6
αβ / ρ
βG((G))
20
CRP activation (α )
Extracellular TMG (μ M)
0.4
0.2
0.0
0 05 0.10
0.05
0 10 0.15
0 15 0.20
0 20
1/ρ
39
40
WT
+7 lacO
+15 lacO
Turning a binary response into
ag
graded response
p
1/ρ = 0.005
Green
flu
uorescence
e
1/ρ = 0.16
20
15
10
αβ/ρ
5
0
no hysteresis
0
100
200
300
400
Extracellular TMG (μM)
41