gaubert et al 08_mol ecol res.doc
advertisement

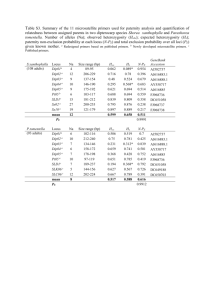
Development and characterization of 11 microsatellite loci in a historically introduced carnivoran, the common genet (Genetta genetta) P. G A U B E R T ,*‡ I . D E L C E R R O ,*§ F. PA L O M A R E S * and J . A . G O D O Y † *Departamento de Biología de la Conservación, †Departamento de Ecología Integrativa, Estación Biológica de Doñana, CSIC, Pabellón del Perú, Avda María Luisa s/n, 41013 Sevilla, Spain, ‡UR IRD 131 — UMS MNHN 403, Département Milieux et Peuplements Aquatiques, Muséum National d’Histoire Naturelle, 43 rue Cuvier, 75005 Paris, France, §Genética de la Conservación Animal, IRTA, Ctra de Cabrils, s/n 08348 Cabrils, Barcelona, Spain Abstract Microsatellite markers were developed to assess population structure and patterns of translocation in the introduced European common genet (Genetta genetta). Primer pairs were designed for 60 microsatellite sequences enriched for CA, GA, CATC and TAGA repeat motifs. Eleven loci that proved to be polymorphic were genotyped in 33 individuals from southwestern France. The number of alleles per locus and observed heterozygosities varied from three to seven and from 0.2121 to 0.7576, respectively. One locus (B103) showed significant departure from Hardy–Weinberg equilibrium, probably due to the presence of null alleles. Tests of linkage disequilibrium did not detect significant associations among loci. Keywords: Carnivora, Europe, Genetta genetta, introduced species, microsatellite, Viverridae Received 13 January 2008; revision accepted 7 April 2008 Genets (Genetta spp.) are nocturnal, solitary, medium-sized predators representing the most speciose carnivoran genus in sub-Saharan Africa (Gaubert et al. 2005). One species, the common genet Genetta genetta (Linnaeus, 1756), also occurs naturally in Maghreb and in the Arabian Peninsula. It was probably introduced from North Africa into southwestern Europe during historical times (Delibes & Gaubert in press), representing one rare example of successful, historical translocation of a wild African mammal into Europe (Gaubert et al. 2008). Thus, the study of the genetic structure and historical demography among its populations may shed light on the population dynamics and short-term evolution at disequilibrium of invading meso-predators. We report herein the 11 first microsatellite markers developed for G. genetta, and assess their level of polymorphism within a French population. Microsatellite isolation was performed by Genetic Identification Services (http://www.genetic-id-services.com) following the magnetic bead-based enrichment procedure of Jones et al. (2000, 2002). Genomic DNA was extracted Correspondence: Philippe Gaubert, Fax: (33)1 40 79 37 71; E-mail: gaubert@mnhn.fr from a muscle sample of a single individual, and was partially restricted with a cocktail of seven blunt-end cutting enzymes (RsaI, HaeIII, Bsr B1, PvuII, StuI, ScaI, and EcoRV). Libraries were prepared in parallel using BiotinCA(15), Biotin-GA(15), Biotin-CATC(8), and Biotin-TAGA(8) (Integrated DNA Technologies) as capture molecules (libraries A, B, C and D, respectively). A total of 100 positive plasmid clones containing inserts 350–700-bp long were sequenced, of which 84 contained a microsatellite (CA, 24/26; GA, 26/28; TAGA, 21/25; CATC, 13/16). Polymerase chain reaction (PCR} primers were designed for 60 microsatellite-containing clones using DesignerPCR version 1.03 (Research Genetics), and PCR was optimized for 26 microsatellites from all four libraries, selecting primers to span different product size ranges. Amplification products were initially labelled fluorescently through an M13–5′-tailed forward primer approach (Boutin-Ganache et al. 2001). These markers were first evaluated for amplification in three tissue samples using ‘touchdown-PCR’. Twenty-four markers rendering clean and efficient amplification of the expected product were evaluated for polymorphism in a set of 11 samples of diverse origin (five from North Africa, four from Spain, one from France and one from Italy). At Table 1 Characterization of 11 microsatellite loci in Genetta genetta among 33 individuals from southwestern France. The letter preceding each locus name refers to the library from which it was selected (see text) Locus Repeated motif GenBank Accession no. Primer sequences (5′–3′) A5 (GT)16 EU362975 A104 (CA)23 EU362976 A108 (CA)16(TGCACACGCACGCG) EU362977 (CA)12 A110 (AC)22 EU362978 A112 (GT)21 EU362979 B103* (GA)26 EU362980 B104 (AG)20 EU362981 B105 (GA)18 EU362982 C101 (ATGG)12 EU362983 D4 (TATC)15 EU362984 D111 (TAGA)14 EU362985 F: GAACTCGGGGCTTAGATGTC R: CTGGAAAGATGAGGGGACTT F: TGAAAGAATTGCTTGGTATGG R: GCATGGTTGGTGAACATTC F: TGCATTACAATCACTCACTCTC R: TAGGTGGAAATCAATCTGTTG F: TCGTGCTGACGTGTTTAGC R: TTTGCCTTCCACAAAGAGG F: CCAACTGCCTCTGTGACTC R: CCAAAACCTATCCGAGAATG F: CTTTCTTGCTGTGTCCTCACA R: GGGTATAGGGCCTTTAAGGAG F: ATCTGCTACTGGCAAGTCAAC R: GCCTGTTTCAGTTTCTGTGTC F: CGTGTATGTGTGTGGTGTGTG R: CCCCTACCTTCTTCATCCAAC F: TCCCACAGAAGGAACAGTC R: GCTTGTCCCATCAGAGTGT F: TTGGAGAGGATTTCACTGAC R: TAGGCTTAGGAGATTTAGCAAG F: TGCTTTTTCTTTAATCCCTCTC R: TATCCTCAGCAGTCCTCAGAG Ta MgCl2 Size range Na HO Label (°C) (mm) (bp) HE FIS FAM 54 2 127–131 3 0.6061 0.6318 0.056 VIC 53 1.5 117–133 4 0.6970 0.5914 –0.164 NED 54 1.5 127–137 5 0.3333 0.5376 0.393 NED 54 2 256–274 5 0.4242 0.4766 0.125 PET 54 2 289–311 4 0.5455 0.5083 –0.058 VIC 60 1 178–190 4 0.2121 0.6331 0.673 VIC 54 1.5 247–261 5 0.5455 0.5106 –0.053 PET 54 2 169–173 2 0.3030 0.2571 –0.164 FAM 54 2 264–276 4 0.3030 0.4187 0.290 PET 2 209–249 7 0.6970 0.7856 0.128 1.8 185–201 5 0.7576 0.7273 –0.026 54 NED 54 F, forward; R, reverse. Label, fluorescent dye used. Ta, annealing temperature. Na, number of alleles. HO, observed heterozygosity. HE, expected heterozygosity. FIS, inbreeding coefficient. *Locus showing departure from Hardy–Weinberg equilibrium (P < 0.05). this stage, we were able to obtain a first estimate of the numbers of alleles and allelic size range for each marker. Twelve markers that showed easily scorable amplification products and high variability were grouped in four nonoverlapping sets that were distributed among the four possible fluorescent labels, so that they could be simultaneously analysed in a single run of capillary electrophoresis. Assessment of polymorphism within 33 individuals from southwestern France was conducted using 5′-fluorescently labelled forward primers and 5′-GTTTCTT-tailed reverse primers (Brownstein et al. 1996). Amplification reactions were performed in 20 μL final volumes consisting of: reaction Buffer 1× (Bioline), 1–2 mm MgCl2 (see Table 1), 0.25 mm dNTPs, 0.25 μm primers, 0.1 mg/mL BSA, 0.025 U/μL Taq DNA polymerase, and c. 50 ng of template DNA. Temperature cycling was performed on MJ Research PTC-100 thermocyclers using the following conditions: initial denaturation of 94 °C for 2 min, followed by 40 cycles of 92 °C (30 s), 53–60 °C (see Table 1) (30 s), and 72 °C (30 s), and terminated with a final extension at 72 °C for 5 min. Products were analysed on an ABI 3100 DNA sequencer and alleles were scored with GeneMapper 4.0 (Applied Biosystems). The genetic diversity of the microsatellite loci was quantified by the number of alleles per locus, observed heterozygosity (HO), and expected heterozygosity (HE) using Genetix version 4.05 (Belkhir et al. 1996–2004). Departures from Hardy–Weinberg equilibrium and linkage disequilibrium for each pair of loci were calculated using permutation tests with fstat version 2.9.3. (Goudet 1995). P values were adjusted for multiple test of significance using the sequential Bonferroni correction at the 5% nominal level (Rice 1989). One locus proved monomorphic for southwestern France — but was polymorphic at a larger geographical scale — and is thus not further discussed here [A113: (GT)5(ATAT)(GT)4(GA)(GT)18); F: 5′-TAAAGGGGTGTGTGTGTATATG-3′ — R: 5′-CCCTATAACACTCTGGCTATTG-3′; annealing temperature: 54 °C; MgCl2: 2 mm; GenBank Accession no. EU362986]. We were able to detect three to seven alleles per locus (average = 4.36). Observed and expected heterozygosities varied from 0.2121 to 0.7576 and from 0.2571 to 0.7856, respectively (Table 1). One locus (B103) showed significant departure from Hardy–Weinberg equilibrium (P = 0.0008) probably due to presence of null alleles. Tests of linkage disequilibrium revealed no significant associations among all pairs of loci. © 2008 The Authors Journal compilation © 2008 Blackwell Publishing Ltd We have identified a set of 11 microsatellites that will be useful for studying population structure, gene flow (natural or following translocations) and historical demography among populations of G. genetta introduced into Europe. An additional locus (A113) will be useful in broader-scale examinations across the entire Mediterranean Basin. This is an important step towards our understanding of the translocation patterns at the origin of the European populations of common genets (Gaubert et al. in press). Acknowledgements We thank Mark Todd and Ken Jones at Genetic Identification Services (Chatsworth, CA) for building the microsatellite library and for fruitful comments on the manuscript. We are grateful to Ana Piriz and Laura Soriano for laboratory work assistance at the Laboratorio de Ecologia Molecular. Pascal and Christine Fournier (Groupe de Recherche et d’Etude pour la Gestion de l’Environnement, Villandraut, France) supplied samples from southwestern France (other field collectors are thanked in Gaubert et al. in press). An anonymous reviewer provided useful comments on the early version of the manuscript. This work was funded by the European Commission 6th PCRDT ‘EPISARS’ (FP6-2003-SSP-2-SARS; no. 51163). References Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F (1996–2004) Genetix 4.05, Logiciel sous Windows pour la Génétique des Populations. Laboratoire Génome, Populations, Interactions, CNRS UMR 5171, Université de Montpellier II, Montpellier, France. http:// www.genetix.univ-montp2.fr/genetix/genetix.htm. Boutin-Ganache I, Raposo M, Raymond M, Deschepper CF (2001) M13-tailed primers improve the readability and usability of microsatellite analyses performed with two different allele-sizing methods. BioTechniques, 31, 25–28. Brownstein MJ, Carpten JD, Smith JR (1996) Modulation of nontemplated nucleotide addition by Taq DNA polymerase: primer modifications that facilitate genotyping. BioTechniques, 20, 1004– 1010. Delibes M, Gaubert P (in press) Genetta genetta Linneaus. In: The Mammals of Africa, Vol. 5. Carnivora, Pholidota, Perissodactyla (eds Kingdon JS, Hoffmann M). Academic Press, Amsterdam, The Netherlands. Gaubert P, Godoy JA, del Cerro I, Palomares F (in press) Early phases of a successful invasion: mitochondrial phylogeography of the common genet (Genetta genetta) within the Mediterranean Basin. Biological Invasions. Gaubert P, Taylor PJ, Veron G (2005) Integrative taxonomy and phylogenetic systematics of the genets (Carnivora, Viverridae, genus Genetta): a new classification of the most speciose carnivoran genus in Africa. In: African Biodiversity: Molecules, Organisms, Ecosystems (eds Huber BA, Sinclair BJ, Lampe K-H), pp. 371–383. Springer, New York. Gaubert P, Jiguet F, Bayle P, Angelici FM (2008) Has the common genet (Genetta genetta) spread into south-eastern France and Italy? Italian Journal of Zoology, 75, 43–57. Goudet J (1995) fstat (version 1.2): a computer program to calculate F-statistics. Journal of Heredity, 86, 485–486. Jones KC, Levine KF, Banks JD (2000) DNA-based genetic markers in black-tailed and mule deer for forensic applications. California Fish and Game, 86, 115–126. Jones KC, Levine KF, Banks JD (2002) Characterization of 11 polymorphic tetranucleotide microsatellites for forensic applications in California elk (Cervus elaphus canadensis). Molecular Ecology Notes, 2, 425– 427. Rice WR (1989) Analysing tables of statistical test. Evolution, 43, 223–225.