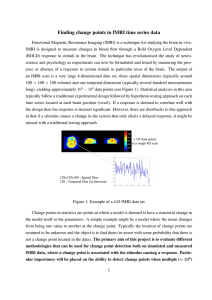
POSTER (PowerPoint)
advertisement
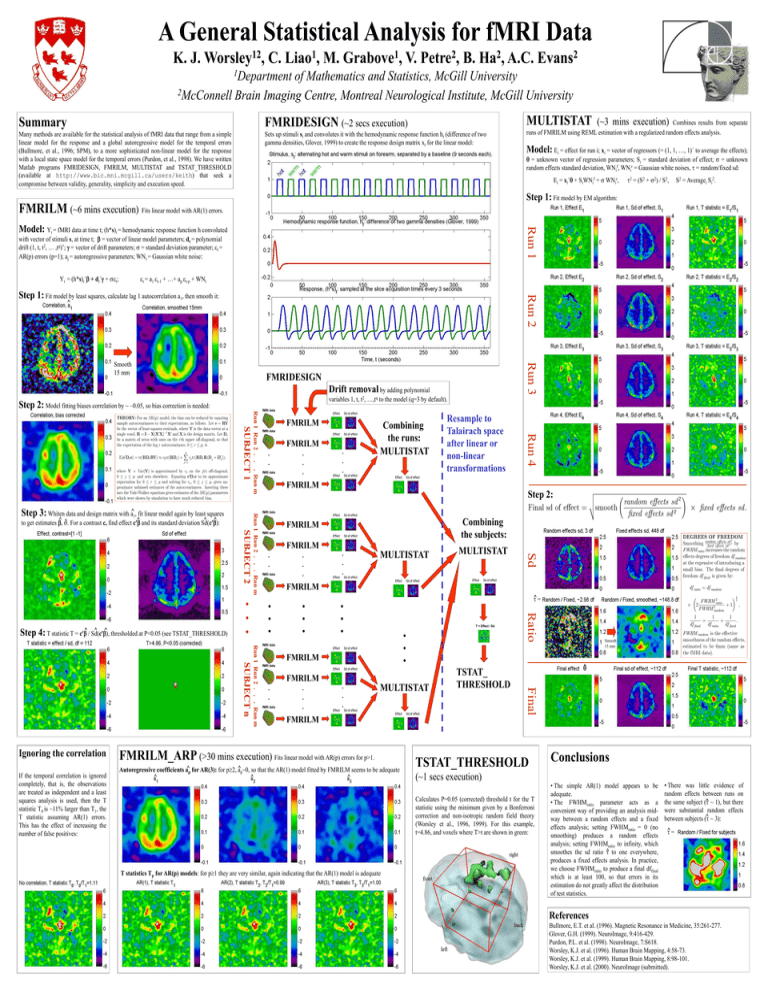
A General Statistical Analysis for fMRI Data K. J. 12 Worsley , C. 1 Liao , M. 1 Grabove , V. 2 Petre , B. 2 Ha , A.C. 2 Evans 1Department of Mathematics and Statistics, McGill University 2McConnell Brain Imaging Centre, Montreal Neurological Institute, McGill University MULTISTAT Summary FMRIDESIGN (~2 secs execution) Many methods are available for the statistical analysis of fMRI data that range from a simple linear model for the response and a global autoregressive model for the temporal errors (Bullmore, et al., 1996; SPM), to a more sophisticated non-linear model for the response with a local state space model for the temporal errors (Purdon, et al., 1998). We have written Matlab programs FMRIDESIGN, FMRILM, MULTISTAT and TSTAT_THRESHOLD (available at http://www.bic.mni.mcgill.ca/users/keith) that seek a compromise between validity, generality, simplicity and execution speed. Sets up stimuli st and convolutes it with the hemodynamic response function ht (difference of two gamma densities, Glover, 1999) to create the response design matrix xt for the linear model: (~3 mins execution) Combines results from separate runs of FMRILM using REML estimation with a regularized random effects analysis. Model: Ei = effect for run i; xi = vector of regressors (= (1, 1, …, 1)´ to average the effects); = unknown vector of regression parameters; Si = standard deviation of effect; = unknown random effects standard deviation, WNif, WNir = Gaussian white noises, = random/fixed sd: Ei = xi´ + SiWNif + WNir, 2 = (S2 + 2) / S2, S2 = Averagei Si2. Step 1: Fit model by EM algorithm: FMRILM (~6 mins execution) Fits linear model with AR(1) errors. Run 1 Model: Yt = fMRI data at time t; (h*s)t = hemodynamic response function h convoluted with vector of stimuli s, at time t; = vector of linear model parameters; dt = polynomial drift (1, t, t2, … ,tq)’; = vector of drift parameters; = standard deviation parameter; t = AR(p) errors (p=1); aj = autoregressive parameters; WNt = Gaussian white noise: Yt = (h*s)t´ + dt´ + t; t = a1 t-1 + …+ ap t-p + WNt Run 2 Step 1: Fit model by least squares, calculate lag 1 autocorrelation a1, then smooth it: ^ Run 3 Smooth 15 mm FMRIDESIGN Drift removal by adding polynomial variables 1, t, t2, …,tq to the model (q=3 by default). Step 2: Model fitting biases correlation by ~ –0.05, so bias correction is needed: fMRI data FMRILM Combining the runs: MULTISTAT FMRILM Step 2: fMRI data Combining the subjects: FMRILM fMRI data fMRI data FMRILM MULTISTAT MULTISTAT FMRILM • • • • ^ = • • • T = Effect / Sd fMRI data FMRILM fMRI data fMRI data FMRILM MULTISTAT TSTAT_ THRESHOLD Smooth 15 mm ^ Final Run 1 Run 2 . . . Run m SUBJECT n • • • Ratio • If the temporal correlation is ignored completely, that is, the observations are treated as independent and a least squares analysis is used, then the T statistic T0 is ~11% larger than T1, the T statistic assuming AR(1) errors. This has the effect of increasing the number of false positives: • • • • Ignoring the correlation fMRI data Sd Run 1 Run 2 . . . Run m SUBJECT 2 ^ ^ thresholded at P<0.05 (see TSTAT_THRESHOLD) Step 4: T statistic T = c^ / Sd(c), FMRILM Resample to Talairach space after linear or non-linear transformations Run 4 . Run m ^ . ^ and its standard deviation Sd(c): ^ ^ ^ For a contrast c, find effect c to get estimates , Run 1 Run 2 . . SUBJECT 1 Step 3: Whiten data and design matrix with a^1, fit linear model again by least squares fMRI data FMRILM FMRILM_ARP (>30 mins execution) Fits linear model with AR(p) errors for p>1. Autoregressive coefficients a^p for AR(3): for p2, ^ ap~0, so that the AR(1) model fitted by FMRILM seems to be adequate ^ ^ ^ TSTAT_THRESHOLD (~1 secs execution) Calculates P=0.05 (corrected) threshold t for the T statistic using the minimum given by a Bonferroni correction and non-isotropic random field theory (Worsley et al., 1996, 1999). For this example, t=4.86, and voxels where T>t are shown in green: T statistics Tp for AR(p) models: for p1 they are very similar, again indicating that the AR(1) model is adequate Conclusions • The simple AR(1) model appears to be adequate. • The FWHMratio parameter acts as a convenient way of providing an analysis midway between a random effects and a fixed effects analysis; setting FWHMratio = 0 (no smoothing) produces a random effects analysis; setting FWHMratio to infinity, which smoothes the sd ratio ^ to one everywhere, produces a fixed effects analysis. In practice, we choose FWHMratio to produce a final dffinal which is at least 100, so that errors in its estimation do not greatly affect the distribution of test statistics. • There was little evidence of random effects between runs on ^ ~ 1), but there the same subject ( were substantial random effects ^ ~ 3): between subjects ( ^ = References Bullmore, E.T. et al. (1996). Magnetic Resonance in Medicine, 35:261-277. Glover, G.H. (1999). NeuroImage, 9:416-429. Purdon, P.L. et al. (1998). NeuroImage, 7:S618. Worsley, K.J. et al. (1996). Human Brain Mapping, 4:58-73. Worsley, K.J. et al. (1999). Human Brain Mapping, 8:98-101. Worsley, K.J. et al. (2000). NeuroImage (submitted).