1 Nucleic Acids and Proteins TOC
advertisement

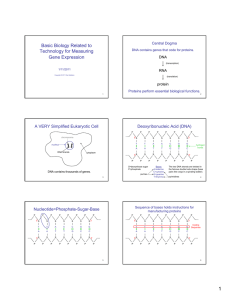
1 Nucleic Acids and Proteins Modern Linguistics for the Genomics and Bioinformatics Era TOC GENOME ↕ CHROMOSOME ↕ GENE ↕ DNA ↕ NUCLEOTIDE Fig. 1. Simple organizational hierarchy of genetic material. GENOME (1) ↕ CHROMOSOME (23) ↕ DNA (23) ↕ GENE (~30,000) ↕ NUCLEOTIDE (3 × 109) Fig. 2. Numerically annotated hierarchy of genetic material. Fig. 3. Molecular structure of nucleotides, the building blocks of DNA and RNA. The 1' Æ 5' numbering convention used to designate carbons in the deoxyribose and ribose sugars is annotated only on the deoxyribose sugar. “B” extending from the 1' carbon of deoxyribo- and ribonucleotides represent the purine and pyrimidine nitrogenous bases. Designation of the different phosphate groups (a, b, g) are depicted on the deoxyribose nucleotide. TOC Fig. 4. Molecular structure of the tetranucleotide GTAG. The H present at 2' carbon of each sugar indicates that this is a DNA chain composed of deoxynucleotides. Phosphates involved in cementing adjacent nucleotides together with a phosphodiester bond are shown in red (on CD). A single free, 5' phosphate is pictured in blue. A free 3'-OH group at the opposite end of the chain is also depicted in blue. Fig. 5. Complementarity between specific nitrogenous bases. Pictured are the A-T and G-C base pairs present in a DNA double helix. (- - - -) represent hydrogen bonds between participating atoms. A purine juxtaposed with a pyrimidine after hydrogen bond formation generates a dimension of 20 angstroms, the width of a DNA double helix. TOC Fig. 6. Base pair complementarity is the basis for faithful duplication of the double helix. Chains of a DNA helix are pictured in an antiparallel configuration. Horizontal lines between the complementing bases denote hydrogen bonds (see Fig. 4). DNA polymerase (oval) polymerizes DNA in a 5' to 3' in an antiparallel direction on each strand. Each new daughter helix is composed of an original stand and a newly synthesized chain, indicative of semi-conservative replication. Fig. 7. A transfer RNA (tRNA) molecule acting as an interpreter between the language of nucleic acids (DNA and RNA) and the language of amino acids (proteins). A tRNA, pictured as a polymer of 76 individual ribonucleotides (individual squares), is folded into the universal cloverleaf structure by virtue of intra-molecular hydrogen bonding between complementing bases. The black dots denote base-pairing by hydrogen bonds to form stems. Gray squares are unpaired nucleotides, forming loops. The triplet anticodon nucleotides (34–36) are shown recognizing and interacting via hydrogen bonding with the appropriate triplet codon within an mRNA molecule. TOC Fig. 8. Molecular structure of the 20 amino acid side-chains (R-groups), listed according to chemical character (A, polar or B, charged). Beneath each structure is the name of the R-group, is threeletter designation, and its single letter designation. In an amino acid, the individual side-chain is attached to by a covalent bond to the common backbone NH2 -CH-COOH, where C indicates the position of attachment. TOC Table 1 Universal Genetic Code First Second Third 5' U C A G 3' U Phe Phe Leu Leu Ser Ser Ser Ser Tyr Tyr Tyr Tyr Cys Cys Stop Trp U C A G C Leu Leu Leu Leu Pro Pro Pro Pro His His Gln Gln Arg Arg Arg Arg U C A G A Ile Ile Ile Met Thr Thr Thr Thr Asn Asn Lys Lys Ser Ser Arg Arg U C A G G Val Val Val Val Ala Ala Ala Ala Asp Asp Glu Glu Gly Gly Gly Gly U C A G Fig. 9. Membrane proteins: An example of how amino acid distribution dictates structure and function. Pictured in black are phospholipids that create the hydrophobic interior of a lipid bi-layer membrane. Small darker dots indicate nonpolar, hydrophobic amino acid constituents of a polypeptide chain. Small lighter dots represent polar, charged, amino acid residues that are capable of interacting with the aqueous, hydrophilic environments on either side of the cell membrane. Such trans-membrane protein domains typify proteins that serve as channels and as receptors to perceive external stimuli. TOC