Genetic & Phenotypic Variation
advertisement
Genetic & Phenotypic
Variation
Genetic variation
• Limits on adaptation
– Natural selection can only work on the variation that is
available through genetic recombination and mutation
DNA and Chromosomes
http://www.accessexcellence.org/RC/VL/GG/chroma_packg.html
Gene Structure
http://genome.imim.es/courses/laCaixa05/Genomes/index.html
Transcription
http://genome.imim.es/courses/laCaixa05/Genomes/index.html
http://www.accessexcellence.org/RC/VL/GG/RNA_trans.html
Exons
http://www.accessexcellence.org/RC/VL/GG/exon2.html
Splicing & Translation
http://genome.imim.es/courses/laCaixa05/Genomes/index.html
Codons
http://www.genome.gov/glossary.cfm#s
Amino Acids and Proteins
http://www.genome.gov/glossary.cfm#s
Mutation
http://www.genome.gov/glossary.cfm#s
Recombination
http://www.blackwell-synergy.com/doi/pdf/10.1111/j.1601-5223.1995.00215.x#search=%22synaptonemal%20complex%20photo%22
http://www.genome.gov/glossary.cfm#s
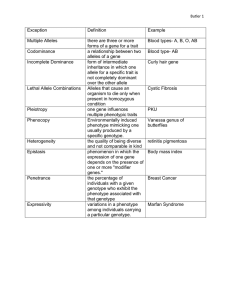
Dominance
http://www.accessexcellence.org/RC/VL/GG/recessive.html
Genotypes
Heterozygous
Homozygous
dominant
Homozygous
recessive
Evolution, Futuyma 2005, Ch.9
Non-coding Genes:
Not such a thing as
Junk DNA
• Non-coding functional RNA
• Cis- and Trans-regulatory
elements
• Introns
• Pseudogenes
• Repeat sequences,
transposons and viral
elements
• Telomeres
Phenotype
Physical or observable characteristics of the individual
Phenotype
• Genotype
• Phenotype
Phenotype
• Genotype
• Environment
– Non genetic
– Env. differences
– Maternal effects
(or paternal)
• Development
• Phenotype
{
•
•
•
•
Temperature, pH, substrate
Yolk composition
Parental care
Plasticity
Phenotype
• Genotype
• Environment
• Development
• Phenotype
• FITNESS
Discontinuous Phenotype
Continuous Phenotype
Threshold traits
Susuki &Nijout 2006, Science
Single Locus Traits
Widow’s peak:
dominant
Variable base pairs
detected by sequencing
Variable sites detected by
restriction enzymes
Attached ear lobe:
recessive
Single nucleotide change at this site
results in AdhS and AdhF alleles
Arrows show short indels
among gene alelles
Mendelian
Inheritance
• Alleles are alternative
forms of a gene located
at one site on a
chromosome (locus);
alleles determine the
traits of individuals.
• Chromosomes and their
alleles separate and
assort independently
when gametes form;
this increases variety
among offspring
Co-dominant alleles
• Genotype predicts the value of the phenotype (red line)
Dominant and recessive alleles
• Genotypic value only imperfectly predicts the value of the
phenotype. Heterozygotes and dominant homozygotes
have the same phenotypic value, thus the fit (red line) has
error (black arrows). Some of the variation is due to additive
genetic causes (red line) and some is due to dominance
variation (black arrows).
Overdominance
• Genotype does not predict the value of the phenotype.
None of the variation is due to "additive genetic effects" of
alleles. All variation is due to error from dominance
variation (black arrows).
Underdominance
• Genotype does not predict the value of the phenotype.
None of the variation is due to "additive genetic effects" of
alleles. All variation is due to error from dominance
variation (black arrows).
Polygenic Traits
•
Controlled by multiple loci (with
incomplete dominance)
AA = dark
Aa = less dark
aa = light
(the same for B
and C loci)
Pleiotropy
•
•
Effect of a single gene on two or more phenotypic
traits
E.g. testosterone controls the development of
secondary sexual characteristics (e.g., a lion's
mane), but also relates to behavioral traits like
aggression.
Epistasis
•
•
Non-additive effect of more than one gene on a
phenotypic trait
It is important in population genetics when the fitness
effects of a genotype depend on what genotype it is
associated with at the other locus.
Multifactorial Traits
• Controlled by >1 loci
(polygenic) +
environment
• Heart disease,
hypertension, diabetes,
obesity, many cancers,
cleft palate (and many
more)
Quantitative Traits
•
•
•
•
•
Continuous traits controlled by multiple loci
with additive effects + environment +
developmental noise
Polygenic
Multifactorial
Numerous possible phenotypic categories
Measurement: continuous, meristic,
threshold
Phenotype
Blue require low pH
P = G + E + (G*E)
P = phenotype of individual
G= genotype of individual
E = environment of individual
G*E = interaction of genotype and environment
Variance Partition
VP = VG + VE
VP = Phenotypic variance within a population
VG = Variance due to genotypic effects
VE = Variance due to environmental effects
Reaction Norms
• Phenotypic range due to environmental effects
• Norms of reactions are often broadest for polygenic characters.
http://www.mun.ca/biology/scarr/4241F_Quantitative_Genetics.html
Phenotypic Plasticity
The ability of a genotype to produce
distinct phenotypes when exposed
to different environments
throughout its ontogeny
Pigliucci 2005, TREE
Miner et al. 2005, TREE
Phenotypic Plasticity
•
Plasticity is therefore a property of the reaction
norm of a genotype, which is the function—
defined in environment / phenotype space—
relating environmental input to phenotypic output
(fig. A, showing three parallel reaction norms).
Vp = Vg + Ve + Vgxe + Verr
•
A: No genetic variation. Plasticity. No GxE.
•
B: Genetic variation. No plasticity. No GxE
•
C: No genetic variation. No plasticity. GxE.
Individual genotypes are different for their
plasticity, but the population as a whole shows no
genetic differentiation (across environment) or
plasticity (across genotypes).
•
D: Genetic variation. Plasticity. GxE
http://www.genotypebyenvironment.org/
Phenotypic Plasticity
Pheidologeton diversus
Pheidole morrisi
Daphnia pulex
Reiber & Roberts 2005, CBP
Sexual Dimorphism
Drosophila melanogaster male (left) & female (right).
Image source: ©www.gen.cam.ac.uk/dept/ashburner.html
Copepods. The male Sappharina only
appears like this when lit at a very specific
angle but is totally transparent under
other lighting conditions. The two sacks
on either side of the female are egg sacks.
Bicyclus anynana
Dicynodonts, a group of four-legged plant eaters,
mammal-like reptiles, thrived from the Late Permian,
around 260 million years ago to the Late Triassic, around
208 million years ago, a time when the Earth's land formed
one giant super-continent, Pangaea.
Sexual Dimorphism
Anastassov et al. 2001, Mol. Biol. Evol.
Epigenetic Variation
• Genomic imprinting
• Chromosomal Inactivation
Epigenetic Modifications
A different layer of trascriptional regulation
Influence gene expression
Includes histone modifications and DNA
methylation
Epigenetic Inheritance
Epigenetic Regulation of Gene Expression
•
•
Epigenetic information
modulates gene
expression without
modifying actual DNA
sequence
Histone modifications
change the chromatin
structure and affect the
accessibility of DNA to
regulatory proteins
•
•
Methylated
DNA
Histone
Gene activation correlated with H3-K9 acetylation
Gene silencing associated with H3-K9 methylation
Phenotype – Development - Fitness