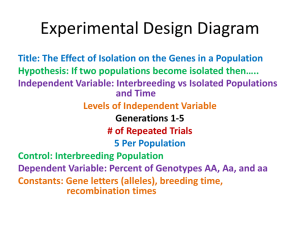
Document 10639601
advertisement

Linear Mixed-Effects Models for Data from Split-Plot Experiments c Copyright 2016 Dan Nettleton (Iowa State University) Statistics 510 1 / 24 An Example Split-Plot Experiment Whole Plot or Main Plot Field Block 1 Genotype C 0 Block 2 100 150 50 Genotype B 150 100 50 0 Genotype A Genotype A 50 100 150 0 Genotype A 0 Genotype B 150 100 0 Genotype C 50 150 100 100 Genotype B 50 50 150 Split Plot or Sub Plot 0 Genotype C Block 3 100 50 0 150 Genotype B 0 100 150 50 Genotype C 50 100 150 0 Genotype A Block 4 0 50 100 150 150 100 c Copyright 2016 Dan Nettleton (Iowa State University) 50 0 50 150 100 0 Statistics 510 2 / 24 This experiment has two factors: genotype and fertilizer amount. Genotype has levels A, B, and C. Fertilizer has levels 0, 50, 100, 150 lbs. N / acre. Genotype is called the whole-plot (or main-plot) factor because its levels are randomly assigned to whole plots (main plots). Fertilizer is called the split-plot factor because its levels are randomly assigned to split plots within each whole plot. c Copyright 2016 Dan Nettleton (Iowa State University) Statistics 510 3 / 24 Experimental Units in Split-Plot Designs Whole plots are the whole-plot experimental units because the levels of the whole-plot factor (genotype) are randomly assigned to whole plots. The split-plots are the split-plot experimental units because the levels of the split-plot factor (amount of fertilizer) are randomly assigned to split plots within each whole plot. Thus, we have two different sizes of experimental units in split-plot experimental designs. c Copyright 2016 Dan Nettleton (Iowa State University) Statistics 510 4 / 24 Same Treatment Structure in an RCBD Field Block 1 B B A C B C A A C B C A 100 0 0 100 150 50 50 150 150 50 0 100 Block 2 A A C A B B C C A C B B 150 0 50 50 100 50 100 0 100 150 150 0 Block 3 C 0 A A B B B A C A C C B 0 100 100 50 0 150 50 50 150 100 150 Block 4 B C B A C A B C B C A A 0 150 50 150 100 0 150 50 100 0 100 50 c Copyright 2016 Dan Nettleton (Iowa State University) Statistics 510 5 / 24 Same Treatment Structure in an CRD Field B B A B A C A B A C C C 50 0 150 100 100 150 50 0 50 100 0 100 A A C B B B A 50 0 50 50 150 50 0 C 0 A A A A 0 100 150 0 C A C B B 0 100 50 150 0 B A B A B C A 0 150 150 50 150100 100 B B B C C C A C C C C B 50 100 100 150 100 50 150 50 150 0 150 100 c Copyright 2016 Dan Nettleton (Iowa State University) Statistics 510 6 / 24 Why Use a Split-Plot Design? Split-plot designs usually arise because logistical constraints make a CRD or RCBD impractical. For example, it may be easier to change from one fertilizer level to another as a tractor drives through a field, while it may be more difficult to change from planting one genotype to planting another. In the engineering literature, split-plot designs are sometimes called designs with hard-to-change factors. c Copyright 2016 Dan Nettleton (Iowa State University) Statistics 510 7 / 24 Recognizing Designs with Split-Plot Structures Many variations on split-plot designs are used for practical reasons. Examples include split-split-plot designs and split-block designs, but the names of these designs are not so important. Pay close attention to the experimental unit to which the levels of each factor are randomly assigned to recognize split-plot-like design structures. c Copyright 2016 Dan Nettleton (Iowa State University) Statistics 510 8 / 24 Split-plot designs may not involve plots of land. Suppose eight pairs of mice from eight litters are housed in eight cages so that each cage holds two mice from the same litter. Suppose diets 1 and 2 are randomly assigned to the litters with four litters per diet. Within each cage, suppose drugs 1 and 2 are randomly assigned to the mice with one mouse per drug. c Copyright 2016 Dan Nettleton (Iowa State University) Statistics 510 9 / 24 A Split-Plot Experimental Design Diet 1 Drug 2 Diet 2 Drug 1 Drug 2 Drug 2 Drug 1 Drug 2 Drug 2 Drug 1 Drug 2 Diet 1 Drug 1 Diet 1 Diet 2 Drug 1 Drug 2 Diet 2 Diet 2 Drug 2 Drug 1 Drug 1 Diet 1 c Copyright 2016 Dan Nettleton (Iowa State University) Drug 1 Statistics 510 10 / 24 Diet is the whole-plot treatment factor. Litters are the whole-plot experiment units. Drug is the split-plot treatment factor. Mice are the split-plot experiment units. c Copyright 2016 Dan Nettleton (Iowa State University) Statistics 510 11 / 24 Diet i = 1, 2, Drug j = 1, 2, Litter k = 1, 2, 3, 4 (within each Diet i) yijk = µ + αi + βj + γij + `ik + eijk (i = 1, 2; j = 1, 2; k = 1, ..., 4) µ + αi + βj + γij = mean for Diet i and Drug j `ik = random effect for kth litter that received Diet i eijk = random error effect for Diet i, Drug j, Litter k c Copyright 2016 Dan Nettleton (Iowa State University) Statistics 510 12 / 24 y111 y121 y112 y122 y113 y123 y114 y124 y= y211 y221 y212 y 222 y213 y223 y 214 y224 µ α1 α 2 β1 β = β2 γ 11 γ12 γ21 c Copyright 2016 Dan Nettleton (Iowa State University) γ22 `11 `12 `13 ` 14 u= `21 `22 ` 23 `24 e111 e121 e112 e122 e113 e123 e114 e124 e= e211 e221 e212 e 222 e213 e223 e 214 e224 Statistics 510 13 / 24 X= 1 , I ⊗ 1, 1 ⊗ I , I ⊗ 1 ⊗ I 16×1 2×2 8×1 8×1 2×2 2×2 4×1 2×2 Z= I ⊗ 1 8×8 2×1 y = Xβ + Zu + e c Copyright 2016 Dan Nettleton (Iowa State University) Statistics 510 14 / 24 " # u " # " # " #! 0 σ`2 I 0 G 0 ∼N , = e 0 0 σe2 I 0 R Var(Zu) = ZGZ0 = σ`2 ZZ0 0 2 = σ` I ⊗ 1 I ⊗ 1 8×8 2×1 8×8 2×1 = σ`2 I ⊗ 11 0 8×8 2×2 " = Block Diagonal with blocks c Copyright 2016 Dan Nettleton (Iowa State University) σ`2 σ`2 # σ`2 σ`2 Statistics 510 15 / 24 0 Var(y) = ZGZ0 + R = σ`2 I ⊗ 11 + σe2 I 8×8 2×2 = Block Diagonal with blocks " σ`2 + σe2 σ`2 σ`2 σ`2 + σe2 c Copyright 2016 Dan Nettleton (Iowa State University) # Statistics 510 16 / 24 Thus, the covariance between two observations from the same litter is σ`2 and the correlation is σ`2 2 σ` +σe2 . These computations can also be done using the non-matrix expression of the model. ∀i, j Var(yijk ) = Var(µ + αi + βj + γij + `ik + eijk ) = Var(`ik + eijk ) = σ`2 + σe2 . c Copyright 2016 Dan Nettleton (Iowa State University) Statistics 510 17 / 24 Cov(yi1k , yi2k ) = Cov(µ + αi + β1 + γi1 + `ik + ei1k , µ + αi + β2 + γi2 + `ik + ei2k ) = Cov(`ik + ei1k , `ik + ei2k ) = Cov(`ik , `ik ) + Cov(`ik , ei2k ) + Cov(ei1k , `ik ) + Cov(ei1k , ei2k ) = Cov(`ik , `ik ) + 0 + 0 + 0 = Var(`ik ) = σ`2 . c Copyright 2016 Dan Nettleton (Iowa State University) Statistics 510 18 / 24 Back to the Traditional Split-Plot Experimental Design Field Block 1 Genotype C 0 Block 2 100 150 50 Genotype B 150 100 50 0 Genotype A Genotype A 50 100 150 0 Genotype A 0 Genotype B 150 100 0 Genotype C 50 150 100 100 Genotype B 50 50 150 0 Genotype C Block 3 100 50 0 150 Genotype B 0 100 150 50 Genotype C 50 100 150 0 Genotype A Block 4 0 50 100 150 150 100 c Copyright 2016 Dan Nettleton (Iowa State University) 50 0 50 150 100 0 Statistics 510 19 / 24 A Model for Data from the Traditional Split-Plot Experiment Genotype i = 1, 2, 3, Fertilizer j = 1, 2, 3, 4, Block k = 1, 2, 3, 4 yijk = µij + bk + wik + eijk µij = mean for Genotype i, Fertilizer j bk = random effect for Block k wik = random effect for Genotype i whole plot in Block k eijk random error effect for Genotype i, Fertilizer j, Block k c Copyright 2016 Dan Nettleton (Iowa State University) Statistics 510 20 / 24 To express the model precisely in vector and matrix form as y = Xβ + Zu + e, we will sort the data first by Block, then Genotype, and then Fertilizer: y = [y111 , y121 , y131 , y141 , y211 , y221 , y231 , y241 , . . . , y314 , y324 , y334 , y344 ]0 e = [e111 , e121 , e131 , e141 , e211 , e221 , e231 , e241 , . . . , e314 , e324 , e334 , e344 ]0 c Copyright 2016 Dan Nettleton (Iowa State University) Statistics 510 21 / 24 X= 1 ⊗ I , 4×1 12×12 c Copyright 2016 Dan Nettleton (Iowa State University) β= µ11 µ12 µ13 µ14 µ21 µ22 µ23 µ24 µ31 µ32 µ33 µ34 Statistics 510 22 / 24 Z= I ⊗ 1 , 4×4 12×1 I ⊗ 1 12×12 4×1 b1 . .. " # b4 b u= = w11 ∼N w w21 . . . " 0 0 # " , #! σb2 I 0 0 σw2 I w34 c Copyright 2016 Dan Nettleton (Iowa State University) Statistics 510 23 / 24 b 0 σb2 I 0 0 w ∼ N 0 , 0 σw2 I 0 e 0 0 0 σe2 I " # " # " #! u 0 G 0 ∼N , e 0 0 R c Copyright 2016 Dan Nettleton (Iowa State University) Statistics 510 24 / 24