3D Bone Microarchitecture Modeling and Fracture Risk Prediction Hui Li Xiaoyi Li
advertisement

3D Bone Microarchitecture Modeling and Fracture Risk
Prediction
Hui Li
∗
Xiaoyi Li
Lawrence Bone
Department of Computer
Science and Engineering,
State University of New York
at Buffalo, USA
Department of Computer
Science and Engineering,
State University of New York
at Buffalo, USA
Department of Orthopedics,
State University of New York
at Buffalo, USA
Department of Orthopedics,
State University of New York
at Buffalo, USA
Department of Pharmaceutical
Sciences,
State University of New York
at Buffalo, USA
Department of Computer
Science and Engineering,
State University of New York
at Buffalo, USA
hli24@buffalo.edu
Cathy Buyea
buyea@buffalo.edu
xiaoyili@buffalo.edu
Murali Ramanathan
murali@buffalo.edu
bone@buffalo.edu
Aidong Zhang
azhang@buffalo.edu
ABSTRACT
Categories and Subject Descriptors
As prevalence and awareness of osteoporosis increase and
treatments of proven efficacy become available, the demand
for management of patients with the disease will also rise. It
calls for innovative research on understanding of osteoporosis and fracture mechanisms, allowing early and more accurate prediction of bone disease progression. The most widely
validated technique for the diagnosis of osteoporosis is Bone
Mineral Density (BMD) measurement based on dual energy
X-ray absorptiometry (DXA). However, a major limitation
of BMD is that it incompletely reflects the variation in bone
strength. In this paper we develop and evaluate a novel
three-dimensional (3D) computational bone framework capable of providing: (1) Spatio-temporal 3D microstructure
bone model; (2) Derived quantitative measures of 3D bone
microarchitecture; (3) Analysis of BMD and bone strength;
and (4) A state-of-the-art probabilistic approach to analyze
bone fracture risk factors including demographic attributes
and life styles. Beyond efficient 3D bone microstructure representation, quantitative assessment is considered not only
for identifying critical elements in bone microstructure, but
also ensuring effective predictioin of bone diseases in advance. The simulation network model of 3D bone microarchitecture and extensive empirical study on fracture risk improve our understanding of bone disease risk arising from
the complex interplay of the human BMD assessment result
with presence of major risk factors.
I.6.3 [Simulation And Modeling]: Applications; I.6.4
[Simulation And Modeling]: Model Validation and Analysis; J.3 [Life and Medical Sciences]: Health; H.2.8 [Database
Management]: Database Applications—Data Mining
∗corresponding author.
Permission to make digital or hard copies of all or part of this work for
personal or classroom use is granted without fee provided that copies are
not made or distributed for profit or commercial advantage and that copies
bear this notice and the full citation on the first page. To copy otherwise, to
republish, to post on servers or to redistribute to lists, requires prior specific
permission and/or a fee.
ACM-BCB’12 October 7-10, 2012, Orlando, FL, USA
Copyright 2012 ACM 978-1-4503-1670-5/12/10 ...$15.00.
Keywords
3D Microstructure Bone Model, Bone Mineral Density, Bone
strength, Osteoporosis, Fracture Risk Factors, Bayesian Network.
1.
INTRODUCTION
Osteoporosis, which is a systemic skeletal disease characterized by low bone mass and micro-architectural deterioration of bone tissue leading to enhanced bone fragility,
has been recognized as an established and well-defined disease that affects more than 25 million people in the United
States. It is estimated that by 2020, one half of all American citizens over age 50 will be at risk for fractures from
osteoporosis and low bone mass. It requires widespread development of facilities for the diagnosis and assessment of
osteoporosis. Traditional thinking on bone’s deterioration
has focused on bone quantity-described by the bone mass
or bone mineral density (BMD)-as a predictor of fracture
risk[10, 4, 8, 16, 29]. The diagnosis of osteoporosis thus centers on assessment of BMD. With the efforts on diagnosis of
osteoporosis, relationships between BMD and fracture risks
have been widely studied[14, 27]. However, low BMD is actually not the sole factor responsible for the fracture risk.
Other factors like bone microarchitecture, demographic attributes and life styles contribute to bone strength, and the
evaluation of them can speed up determination of bone quality and strength[13, 11, 24, 15].
In this paper, we begin by introducing 3D bone microstructure modeling and validation methodology. Although bone
is a simple composite of a mineral phase which is a calcium phosphate-based hydroxyapatite embedded in an organic matrix of collagen protein, its structure is highly complex. So we identify the important components and properties of bone microstructural units to develop a computa-
Table 1: Common Risk Factors for Osteoporosis
Non-modifiable
Modifiable
Low levels of
Older age
calcium and vitamin D
Reduced intake of
Female gender
vegetables and fruits
Family history
Sedentary lifestyle
Small frame or
Smoking
low body weight
History of broken
Increased use of alcohol
bones or height loss
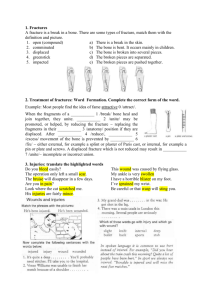
Figure 1: From the bone microstructure, a 3D rodlike structure network is generated as an abstracted
bone network for both normal bone and osteoporotic
bone.
our understanding of bone disease risk.
2.
tional model which reflects the density for mineralized fibers
of bone microstructure. Based on our preliminary study of
the 2D bone structure simulation [18, 19, 20, 21, 22, 23],
we build a 3D bone microarchitecture network. In Figure 1, we show important elements of bone structure and
the abstracted bone model, a rod-like structure network for
both normal and osteoporotic bone. Attributes of bone microarchitecture are considered and applied to our 3D bone
network. Importantly, there is a densely connected region
resembling cortical bone around the outer periphery and a
more loosely connected, less dense region resembling trabecular bone toward the center. We also develop our effective
measurements to quantitatively assess bone quality from biological, mechanical and geometrical perspectives.
Once our 3D bone microstructure model is constructed
and validated, we seek to improve our understanding of major osteoporotic fracture and hip fracture development. As
we know, low BMD is not the only factor that contributes
to the fracture risk. People find a lot of risk factors that are
related to bone fracture which mainly contain demographic
attributes like age, gender and human life styles such as
smoking and excessive alcohol intake. So evaluation of these
risk factors is an important component in the assessment of
bone health. Many falls are preventable, and thus reducing osteoporotic fractures and hip fractures rates and even
morbidity and mortality associated with them. The leading
causes of osteoporosis are separated into non-modifiable and
modifiable risk factors in Table 1. Typically, fracture risk
factors including demographic attributes and life styles for
the fracture prediction are identified using traditional rulebased methods. A widely used FRAX tool developed by
the WHO (World Health Organization) [1] gives the 10-year
probability of fracture risk based on a number of factors
which are input into the calculation. Some papers examining the clinical FRAX score alone can accurately predict
the risk of osteoporotic fracture, and thereby reduce the need
for DXA scans and unnecessary anti-osteoporosis treatment
[12]. It is a “black box” and impossible to mine the relationship between its questions and results. So for diagnosis,
FRAX is convenient to predict the fracture risk in 10 years.
But for analyzing and understanding fracture risk factors, it
is not a good descriptive and flexible tool. Starting from analyzing FRAX tool, the latter part of this paper specializes
in capturing known statistical relationships between fracture
risk factors using probabilistic models and thus improving
METHODS
This section describes our methodology for 3D bone microstructure modeling and measurement, as well as the analysis of bone fracture risk factors.
2.1
Distribution of Bone-Forming Foci and Bone
Mineral
The mean concentration of bone-forming foci c(r) is expressed as the mean number of foci per unit area varied
in the radial direction r according to a Cumulative Normal
Distribution (CDF):
Z r −(z−µ)2
c0
e 2σ2 dz ,
(1)
c(r) = √
2π −r
where the mean and standard deviations of the underlying
normal distributions are denoted by µ and σ, respectively,
and c0 is a constant of proportionality. We have assessed
the ability of this equation to describe data by comparing
with bone imaging. The values of µ and σ can be estimated
by the Nelder-Mead algorithm [26].
The edges of the Pruned Voronoi Lattice (PVL) are assigned weights that are interpreted as the circular crosssectional area of the bone mineral in the edge. The distributions of the circular cross-sectional area of bone mineral
w(r) vary in the radial direction r according to a CDF:
Z r −(z−µ)2
w0
w(r) = √
(2)
e 2σ2 dz ,
2π −r
where the mean and standard deviations of the bone mineral
normal distribution, denoted by µ and σ, respectively, are
fixed at the values used for the distribution of bone forming
foci, and w0 is a constant of proportionality.
2.2
Voronoi Tessellation and Edge Pruning
Voronoi tessellation is a mathematical procedure for creating a closed lattice of edges called Voronoi cells from a set
of points or Voronoi sites [5].
We first generate a field of vertexes as the center of the
bone mineral in the network model. The number of vertexes
per unit area from the center to the boundary varies in the
radial direction according to a CDF:
Z −r −(z−µ)2
λ0
e 2σ2 dz ,
λ(r) = √
(3)
2π −∞
where the mean and standard deviation of the underlying
normal distribution denoted by µ and σ are the same as
those for the concentration on bone mineral in the continuum model, and λ0 is a constant of proportionality.
The point distribution is converted to a network using
the Voronoi tessellation method [7]. Voronoi tessellation
converts a set of points into discrete regions containing of
points that are closer to that point than any other points.
The boundaries of the discrete polygonal Voronoi regions
are straight lines; every point within a given Voronoi cell is
closer to its cognate Voronoi site than to any other site. The
field of bone forming foci created by Equation 1 provides the
set of Voronoi sites that are subject to Voronoi tessellation.
The edges of Voronoi cells are interpreted as the structural
matrix upon which bone mineral is laid down.
Edges in the Voronoi lattice are randomly deleted with
a low probability p according to a Bernoulli distribution
Bernoulli(p). This creates a lattice with characteristics that
more closely approximate observed bone mineral lattices in
bone histological specimens. We refer to the lattice remaining after the stochastic edge-pruning step as the PVL as
defined earlier. The probability of deletion is an exponential function of the edge weight wi , which represents the level
of bone mineral:
p=e
−βwi
wmax
,
(4)
where wmax is the maximum value of wi and β is a nondimensional parameter akin to a rate constant that determines the extent of edge pruning.
2.3 Expression for Bone Mineral
The mean mass of bone mineral m for a given realization
of the network model is obtained as the sum over all the
edges:
X
m=ρ
wi l i ,
(5)
where wi denotes the cross-sectional area of the edge and li
is the length of the edge. The true density of bone mineral
is denoted by ρ.
A BMD test measures the density of minerals in people’s
bones, which is the one most important component estimating the strength of bones. Traditionally, the BMD equation
is:
BM D = BM C/W ,
(6)
where BMC stands for bone mineral content in the unit of
g/cm and W stands for the projected area from DXA scan.
We develop a bone quality measurement using our 3D
bone structure network model of bone dynamics. With
the view of three dimensions, the bone model consists of
nodes and edges which are represented by microscopic sphere
shapes and microscopic cylinder shapes, respectively. Because the model inherits the nature of the network dynamic
modeling properties, we can calculate the structural strength
and network density of the bone model. The method gives
us the measurements that possibly analyze the condition of
the bone model which reflects the variation in bone strength
and bone microstructure. Based on this model, we are able
to assess the bone strength by the following computational
BMD formula:
P
ρ e wi l i
m
m
BM D =
=
,
(7)
=
Ap
πR02
πR02
where m is the mean mass of bone mineral given by Equation
5 and Ap is the projected area of the bone segment, and the
radius of projected area is denoted by R0 .
2.4
Expression for Area Moment
The area moment Iz is a topological property which is an
important determinant of the stress-strain relationships in
response to forces that cause bone to bend.
The area moment is computed relative to the central zaxis of the cylindrical geometry. To compute the area moment Iz due to bone mineral for a realization of the network,
the region r (0 ≤ r ≤ R0 ) is sub-divided into small, ideally
infinitesimal, annular regions and the area δai (r) of the mineral present at distance r from the axis are computed. The
area moment of inertia is defined as the summation over all
the infinitesimal elements in the geometry:
Iz =
X
r2 δai (r) ,
(8)
i
where the value of the radius of gyration of the bone Rg2
is obtained by dividing the area moment Iz by the crosssectional area A. For an circular section of bone:
Rg2 =
2.5
Iz
Iz
=
.
A
πR02
(9)
Bayesian Networks
After constructing the 3D model, we provide novel insights into understanding of a number of risk factors which
are linked to the bone deterioration and may result in an individual’s likelihood of developing the bone fracture. Probabilistic graphical models (PGMs) are a marriage between the
probability theories and graph theories that includes both
directed graphical model and undirected graphical model.
PGMs can be used to model complex probabilistic domain.
One important task for PGMs is to find the assignment to
each variable that jointly maximizes the probability defined
by the model.
Bayesian networks (BNs) are a directed graphical model.
The core of the Bayesian network (BN) representation is a
directed acyclic graph (DAG) G whose nodes represent the
random variables X1 ... Xn and edges denote dependencies. Conditional Probability Distribution (CPD) for each
node Xi describes P (Xi ) | P a(Xi ), where the probability
for each parent node of Xi is denoted by P a(Xi ). BN represents
a joint distribution via the chain rule: P (X1 ...Xn ) =
Q
i P (Xi ) | P a(Xi ).
As a descriptive model, BN provides a complex joint distribution over a large number of random variables and each
variable is directly influenced by only a few other variables.
Meanwhile, it is also a flexible model to answer queries about
causal reasoning, answer queries about diagnostic reasoning, and answer queries about arbitrary mixtures of the two
forms of reasoning.
By modeling an entire joint distribution, BNs can answer
any probability query, in particular, any query where we
want to find the probability distribution over some variables
given evidence about any other variables. In theory, doing inference in BNs is an NP-hard problem. However, the
dependency structure from network representation can be
exploited by inference algorithms. Also, there are some advanced inference algorithms which can handle with querying very large networks. Furthermore, the graph structure
can assist us in inducing cause and effect, and thus understanding and reaching conclusions about the consequences
of various risk factors in our specific domain.
3. MODELING OF BONE MICROSTRUCTURE
In this section, we first show the process of 3D bone network modeling in details and realizations of the bone model
on DXA scan images. Since DXA scan is broadly used to
calculate BMD, important properties of bone strength can
be studied by analyzing bone density distribution and BMD
of DXA scan images [28, 6]. We use a human femur bone
image of DXA scan to analyze properties of the bone density
distribution and BMD.
Figure 2 shows the process of DXA scan and results of
scan images. From the DXA scan, x-ray images and BMD
can be obtained and used as inputs of our mathematical
bone network model. Figure 2(c) shows a femur bone image
of a patient with osteoporosis by DXA scanning with which
we can identify the density distribution of the femur bone.
We can calculate the bone density distribution to the bone
on the DXA scan images of two patients shown in Figure
2(f) [18, 19, 20, 21, 22, 23].
portant factor when the degree of bone strength by various
forces is the major objective of the measurement. The length
and thickness of the bone should be carefully measured before the bone microarchitecture model is created. The general procedure for obtaining a 3D model which represents the
bone architecture contains: (1) generating thousands of twodimensional bone random seeds based on CDF and distributing 3D points with varying radial density according to the
CDF in a circular region in Figure 3(a),(b), (2) implementing Voronoi tessellation on 3D distributing points in Figure
3(c), (3) randomly pruning of extended to infinity Voronoi
cells in Figure 3(e) after finding extreme vertices which are
points on the boundary in Figure 3(d), (4) constructing 3D
bone model surface using triangle representation in Figure
3(f), and (5) constructing Normal Bone Network (NBN) and
Osteoporotic Bone Network (OBN) by pruning edges. The
NBN and OBN construction results from front and top view
are shown as Figure 4(a) and (b).
Figure 2: (a) A DXA machine. (b) A DXA scan image
for whole human body. (c) The femur bone as the major
component to measure BMD. (d) The magnified image
of femur bone. (e) The rotation image of femur bone.
(f ) The density distribution on the DXA scan images.
From BoneNET [23], parameters µ and σ are estimated
by minimizing the squared difference between the density
distribution from a DXA scan image and the density distribution of the mathematical network bone model. By fitting
the parameter values of the model to the density distribution of DXA scan image, the model can realistically reflect
the mineral density in bone microstructure which is an important component of bone strength. Based on preliminary
parameter study, we show the process of our 3D bone modeling in details and realizations of the bone model on DXA
scan images.
In our previous research, one horizontal plane of our 3D
bone microarchitecture model has been studied, which results in a 2D bone structure network [18, 19, 20, 21, 22, 23].
In our 3D bone network model, important components of
the bone structure are considered to build a bone model enabling computational analysis in a timely manner without
losing the critical bone structure data. By stacking up hundreds layered two dimensional bone structure networks, we
create a 3D bone structural network shown in Figure 3. The
number of layers used in this model can be determined by
the research purposes using the model. For example, when a
measurement of quantitative BMD is conducted, the number
of layers could be less than fifty for reducing computation
time on the model. However, the number of layers is an im-
Figure 3: 3D Bone Microstructure Modeling.
Figure 4: (a)NBN front view and top view, (b)OBN
front view and top view.
4. UNDERSTANDING FRACTURE RISK FACTORS USING PGMS
4.1 Structured Graphical Models
We extract 11 attributes including questions and prediction results in the FRAX tool [1] to construct variables
for our Bayesian network in Figure 5. This Bayesian network structure has been designed and analyzed under the
help of UB(University at Buffalo) Orthopaedics and Sports
Medicine Erie County Medical Center. This is an example indicating the interactions between fracture risk factors
(age, sex, body mass index, current smoking, extra alcohol
intake, previous fracture, rheumatoid arthritis, secondary
osteoporosis, parent fractured hip) and fracture risks (osteoporotic fracture, hip fracture). It provides a statistical
model that can unveil and explore many interesting probabilistic dependencies that hold in this domain. For example,
Figure 5 shows three potential evidences, osteoporosis, previous fracture and parent fractured hip, either of which may
cause a patient to have hip fracture. Thus it can be used
for answering queries about any aspect of the domain given
any set of observations.
Figure 5: Fracture risk factors Bayesian network.
We then explore a direct inference method to answer queries
such as P (Xi ) | P a(Xi ). The main steps for this inference method include: (1) fixing a network to prepare for
inference; (2) conditioning on the evidence; (3) computing
the joint distribution and marginalizing out the non-query
nodes efficiently; and (4) performing queries on the bone
fracture risk BNs. Several observations from bone fracture
risk BNs merit further fracture risk investigation and prediction. Figure 6 shows a simple Bayesian network with 7
attributes, together with the conditional probability tables,
representing the dependency structure among the variables.
Figure 6: The CPD for each factor in Bayesian Network.
Table 2: Age Interval
Age
Index
50-54
0
55-59
1
60-64
2
65-69
3
70-74
4
75-79
5
80-84
6
85-89
7
90-95
8
entries as the testing data Xtest = {xl+1 ...xn }. The task is
to predict the labels of the testing data.
We collect the Fracture Risk dataset from the FRAX tool,
which contains about 2000 instances and 11 observed variables. All patients are women in the age interval of 55 to
95. The patients’ questionnaire result includes osteoporotic
fracture risk percentage and hip fracture risk percentage as
shown in the FRAX tool. We also prefer datasets that have
few or no continuous features to avoid information loss. We
present an appropriate method to discretize multivariate
continuous data as follows. We focus on women and men
over age 50 and so we divide age interval by index from 0
in Table 2. Body mass index (BMI kg/m2 ) computed from
height and weight can be regarded as a risk factor. Since low
BMI will lead to an increased risk of fracture, we give the
following definition and transfer to binary value to present
normal or low BMI from weight and height. Normal BMI
= 25.0 kg/m2 is based on average height of 163 cm (64.17
in) and weight of 66.5 kg (146.61 lb). And low BMI = 21.2
kg/m2 is based on the average height of 163 cm (64.17 in)
and weight of 56.7 kg (125 lb) in Table 3. Osteoporotic frac-
4.2 Problem Statement and Dataset
Suppose an instance space X = {x1 ...xl , xl+1 ...xn }, and
label space Y = {0, 1, 2} for 3-class corresponding to low
risk(L), medium risk(M) and high risk(H) for both major
osteoporotic fracture risk and hip fracture risk. We take l
entries as the training data Xtrain = {x1 ...xl } and the rest
Table 3: BMI Interval
BMI
Index
greater than 21.2
0
less than or equal to 21.2
1
Table 4: P(OST|A=3,BMI=1,CS=1,AL=1,PFH=1,HIP=1)
Risk Level Osteoporotic Fracture
2
0.9159
1
0.9
Accuracy
0.8
Table 5: P(HIP|A=3,BMI=1,CS=1,AL=1,PFH=1,OST=1)
Risk Level Hip Fracture
2
1
0.7
0.6
0.5
0.4
ture risk percentage and hip fracture risk percentage should
also be transformed into low, medium and high risk level
based on the guideline provided in [17].
0.3
0.2
OsteoporosisFracture
HipFracture
1
2
3
4
5
6
Number of observed variables
4.3 Answering Queries
We now wish to use our BNs to answer probability queries
about unknown individuals. Table 4 and Table 5 show examples of estimating the probability of osteoporotic fracture (OST) and hip fracture risk (HIP) in US Caucasian
white women, given the observations from the following factors: age(A), sex(S), body mass index(BMI), current smoking(CS), extra alcohol intake(AL), previous fracture(PF),
rheumatoid arthritis(RA), secondary osteoporosis(SO) and
parent fractured hip(PFH). We note that all of the random
variables except age and fracture risk are binary, where the
value “1” represents positive and “0” otherwise. For example, the patient is in the age bucket No.3 (65 to 69 years
old from Table 2) and other factors are all positive, then
our BNs return estimations that the fracture risk is high
for both fracture risk cases (the value of prediction “2” has
the largest possibility which corresponds to high risk level
defined by Problem Statement in Section 4.2).
4.4 Classification Framework
For the fixed directed graphical model as shown in Figure
5, we assume that the age is the root node in the network.
The learning of each node is done by computing the full
joint distribution over the variables in a network, and then
marginalizing out irrelevant variables. We use rudimentary
inference engine for BNs. The classification of each case in
the test set is done by choosing, as class label, the value
of class variable that has the highest posterior probability,
given the instantiations of the feature nodes. The classification accuracy is defined as the percentage of correct predictions on the test sets using a 0-1 loss function that measures
the full penalty for wrong decisions and no penalty otherwise.
The accuracy on each number of observed variables is an
average result. For example, we randomly choose three variables from six given variables in total if only three of them
are observed. We then repeat 20 times. Figure 7 showes that
predicting hip fracture is tougher than osteoporotic fracture,
especially when only a limited number of clinical risk factors
are found. Furthermore, patients do not need to provide all
evidence to predict their fracture risk, while the accuracy
increases as the number of known variables increase. As
we expect in Figure 7, the more you know about a patient’s
risk factors, the more accurately you can predict the fracture
risk.
5. MEASUREMENTS OF BONE QUALITY
In this section, we design mathematical measurements of
Figure 7: Classification result on fracture risk given
different number of observed variables.
Table 6: Moment of Inertia for
Bone Network BMD (g/cm2 )
NBN
374.38
OBN
146.18
NBN and OBN
MOI (g · cm2 )
8.0914e+006
3.8735e+006
bone quality from biological, mechanical and geometrical
perspectives. And we validate both bone networks NBN
and OBN as shown in Figure 4 using these measurements.
5.1
Measuring Bone Strength Based on BMD
and MOI
A projected area density, referred to as BMD, is normally
calculated to assess regional bone density and strength defined by Equation 7. We investigated the effect of BMD on
both bone networks NBN and OBN in Figure 4. We manually set up BMD to a unit by BMD test result according
to the WHO (World Health Organization) T-scores report.
For both NBN and OBN, we solve bone mineral density coefficient ρ in Equation 7. The larger ρ is, the denser bone
network, and vice versa.
We also evaluate the overall strength of the bone network
with moment of inertia (MOI), which is a measure of an
object’s resistance to changes in its rotation rate. Since the
nature of bone strength is the resistance to changes from
outer forces, MOI is a useful measurement for understanding of bone strength. Equation 8 defines area moment, and
the area δai (r) can be regarded as the cross-sectional area
of edge given by Equation 7, because it defines the BMD.
Equation 9 is modified to include a wi term which is the
weight of an edge and the dot product between the direction of the edge ~k as well as the orientation of cross section
~
di.
X wi Ri2
X wi Ri2
,
(10)
M OI =
=
cosθ
d~i~k
e
e
where Ri is the distance from projected area center to object
center.
From Table 6, we can intuitively find that NBN is healthier than OBN because NBN is denser and has better resistance from outer force than OBN.
5.2
Measuring Bone Cross-Sectional Geometry
Figure 8: (a) DXA image fractal analysis. (b) Model projection image fractal analysis.
6.
Fractals are geometric structures that can be used to analyze our trabecular bone structures [2, 25, 9]. Most people are familiar with the topological dimensions such as 1-,
2-, or 3-spatial-dimensions. However, some geometrical objects couldn’t be described well with the usual topological
dimensions and therefore fractional or fractal dimension is
introduced, existing somewhere between the usual topological dimensions [2]. Trabecular bone is a special geometrical
object and can be regarded as a hollow bone or solid bone if
fractal index is 1.0 or 2.0. Measurements of normal bone or
osteoporotic bone usually fall somewhere between these two
extremes and so fractal analysis is useful in estimating bone
strength of different conditions. 2D box-counting algorithm
[3] is applied to calculate the fractal index of trabecular bone
on real DXA image and bone network cross-sectional image.
Fractal dimension results are the negative value of the slope
of the line relating the natural logarithms of the number of
boxes containing contour and each corresponding box size.
We match our NBN and OBN to normal bone trabecular
architecture and osteoporotic bone trabecular architecture
by fractal dimension calibration as shown in Figure 8 using ImageJ tools [9]. Figure 8 left part presents the result
of DXA image fractal analysis. We obtain DXA image for
both healthy bone and osteoporotic bone and then calculate
fractal dimension on both case. Then we process our 3D
model part. We first project our 3D model to a compressed
2D image and then calculate the fractal dimension as shown
in the right part of Figure 8. The objective is to match the
real image fractal dimension with our 3D bone model. The
fractal dimension for real normal trabecular bone image is
1.7273, where the similarity fractal dimension obtained using
box-counting algorithm on our NBN is 1.7993. The fractal
dimension for real osteoporotic bone image is 1.6173, where
the similarity fractal dimension obtained using box-counting
algorithm on our OBN is 1.6086.
We note that the success of matching fractal dimension
is the necessary and startup step for mechanic evaluation
research on our 3D bone network and mechanistic understanding of bone microarchitecture and identifying topological network characteristics related to crack propagation
characteristics and fracture risk.
CONCLUSIONS AND FUTURE WORKS
In this paper, we first developed and evaluated a novel 3D
computational bone framework, which is capable of enabling
quantitative assessment of bone micro-architecture, bone
mineral density and bone’s deterioration. We simulated and
analyzed the microstructure of human normal bone network
and osteoporotic bone network with different bone loss rate.
We then sought to learn relationships between surveyed/selfreported life styles and predicted bone loss rate determined
by 3D bone remodeling. We used Bayesian models to analyze bone fracture risk based on demographic characteristics
and life styles. This probabilistic graphical model can also
answer fundamental questions about fracture risk such as:
Are certain life styles more harmful than others? What are
the most influential factors to bone fracture deterioration?
As a result, people might start to cease drinking alcohol and
smoking to protect their bone health. Finally, we designed
the measurements in biological, mechanical and geometrical domains to validate our model. The validation results
clearly demonstrated our 3D bone model is robust to reflect
the properties of the bone microstructure in the real world.
In the future work, we are more interested in a bone-like
model representing the arbitrary shape of a real bone sample, which might provide more details such as soft material
and hard material in bone microstructure. Moreover, we
will try to learn a dependency structure of a Bayesian network automatically if we can collect heterogeneous dataset
from distributed sites. For the validation part, we will measure our bone model from some mathematical analysis of
bending and buckling parameters of bone structure based
on our current mechanical validation approach. Ultimately,
this study would provide new insights to the understanding
and prediction of bone diseases, such as osteoporosis.
7.
REFERENCES
[1] http://www.shef.ac.uk/FRAX/.
[2] http://www.fractal.org/.
[3] T. G. Smith, Jr., G. D. Lange and W. B. Marks.
Fractal methods and results in cellular morphology.
Journal of Neuroscience Methods, 69:1123–126, 1996.
[4] Ammann, P. and Rizzoli, R. Bone strength and its
determinants. Osteoporos Int, 14:13–18, 2003.
[5] Aurenhammer, F. Voronoi diagrams - a survey of a
fundamental geometric data structure. ACM
Computing Surveys, 23:345–405, 1991.
[6] Barnes, C., Newall, F., Ignjatovic, V., Wong, P.,
Cameron, F., Jones, G., and Monagle, P. Reduced
bone density in children on long-term warfarin.
Pediatric Research, 57:578–581, 2005.
[7] Bock, M., Tyagi, A.K., Kreft, J.U., and Alt, W.
Generalized voronoi tessellation as a model of
two-dimensional cell tissue dynamics. Biological
Physics, 0901:4469, 2009.
[8] Cummings, S.R., Nevitt, M.C., Browner, W.S., Stone,
K., Fox, K.M., Ensrud, K.E., Cauley, J., Black, D.,
and Vogt, T.M. Risk factors for hip fracture in white
women. Study of Osteoporotic fractures research group,
332:767–773, 1995.
[9] Doube M, Klosowski MM, Arganda-Carreras I,
Cordelieres F, Dougherty RP, Jackson J, Schmid B,
Hutchinson JR, Shefelbine SJ. Bonej: free and
extensible bone image analysis in imagej. Bone,
47:1076–1079, 2010.
[10] European Foundation for Osteoporosis. Consensus
development conference: Prophylaxis and treatment of
osteoporosis. Osteonorosis International, 1:114–117,
1991.
[11] Garnero, P., Delmas, P.D. Noninvasive techniques for
assessing skeletal changes in inflammatory arthritis:
bone biomarkers. Curr Opin Rheumatol., 16:428–434,
2004.
[12] Goodhand, J. R. and Kamperidis, N. and Nguyen, H.
and Wahed, M. and Rampton, D. S. Application of
the who fracture risk assessment tool (frax) to predict
need for dexa scanning and treatment in patients with
inflammatory bowel disease at risk of osteoporosis.
Alimentary Pharmacology and Therapeutics,
33(5):551–558, 2011.
[13] Hui, S.L., Slemenda, C.W. and Johnston, C.C. Age
and bone mass as predictors of fracture in a
prospective study. The Journal of Clinical
Investigation, 81:1804–1809, 1988.
[14] Johnell, O., Kanis, J.A., Oden, A., Johansson, H., De
Laet, C., Delmas, P., Eisman, J.A., Fujiwara, S.,
Kroger, H., Mellstrom, D., Meunier, P.J., Melton, L.J.
3rd, O’Neill, T., Pols, H., Reeve, J., Silman, A., and
Tenenhouse, A. Predictive value of bmd for hip and
other fractures. Journal of bone and mineral research,
20:1185–94, 2005.
[15] Johnell, Olof and Nilsson, Bo. Life-style and bone
mineral mass in perimenopausal women. Calcified
Tissue International, 36:354–356, 1984.
[16] Kanis, J.A., Borgstrom, F., De Laet, C., Johansson,
H., Johnell, O., Jonsson, B., Oden, A., Zethraeus, N.,
Pfleger, B., and Khaltaev, N. Assessment of fracture
risk. Osteoporos Int, 164:581–589, 2005.
[17] Kanis JA, Oden A, Johansson H, Borgstrom F, Strom
O, McCloskey E. Frax and its applications to clinical
practice. Bone., 44:734–743, 2009.
[18] Kim, T., Bone, L., Ramanathan, M. and Zhang, A.
Mathematical network model for bone mineral density
(bmd) and bone quality assessment. In the ACM
Conference on Bioinformatics, Computational Biology
and Biomedicine (ACM BCB), pages 69–75, 2011.
[19] Kim, T., Hwang, W., Ramanathan, M., and Zhang, A.
Computational framework for microstructural bone
dynamics model and its evaluation. In IEEE
International Conference on Bioinformatics and
Bioengineering (BIBE), pages 92–98, 2010.
[20] Kim, T., Hwang, W., Zhang, A., Ramanathan, M. and
Sen, S. Damage isolation via strategic self-destruction:
A case study in 2d random networks. Europhysics
Letters, 2:77, 2009.
[21] Kim, T., Koh, J., Li, K., Ramanathan, M. and Zhang,
A. Identification of critical location on a
microstructural bone network. In IEEE International
Conference on Bioinformatics and Biomedicine
(BIBM), pages 557–562, 2010.
[22] Kim, T., Ramanathan, M. and Zhang, A. A
graph-based approach for computational model of
bone microstructure. In ACM Workshop on IWBNA,
in conjunction with ACM International Conference
Bioinformatics and Computational Biology
(ACM-BCB), pages 563–568, 2010.
[23] Kim, T., Ramanathan, M., and Zhang, A. Bonenet: A
network model of bone microstructure and dynamics.
International Journal of Data Mining and
Bioinformatics (IJDMB), accepted.
[24] Krall, Elizabeth A. and Dawson-Hughes, Bess.
Heritable and life-style determinants of bone mineral
density. Journal of Bone and Mineral Research,
8(1):1–9, 1993.
[25] L. Pothuaud, C. L. Benhamou, P. Porion, E.
Lespessailles, R. Harba, P. Levitz M.D. Fractal
dimension of trabecular bone projection texture is
related to three-dimensional microarchitecture.
Journal of Bone and Mineral Research, 15:691–696,
2000.
[26] Nelder, J.A. and R. Mead. A simplex method for
function minimization. Computer Journal, 7:308–313,
1965.
[27] OTT, S.M., Kilcoyne, R.F., and Chesnut, C.H. 3rd.
Ability of four different techniques of measuring bone
mass to diagnose vertebral fractures in
postmenopausal women. Bone Miner Res, 2:201–210,
1987.
[28] Pludowski, P., Lebiedowski, M., and Lorenc, R.S.
Dual energy x-ray absorptiometry. Osteoporos Int,
14:317–322, 2004.
[29] Taylor, B.C., Schreiner, P.J., Stone, K.L., Fink, H.A.,
Cummings, S.R., Nevitt, M.C., Bowman, P.J., and
Ensrud, K.E. Long-term prediction of incident hip
fracture risk in elderly white women: study of
osteoporotic fractures. J Am Geriatr Soc Int,
52:1479–1486, 2004.