Protein interaction network
advertisement

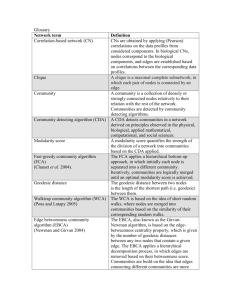
Biological networks Bing Zhang Department of Biomedical Informatics Vanderbilt University bing.zhang@vanderbilt.edu Protein-protein interaction (PPI) Definition Physical association of two or more protein molecules RNA polymerase II, 12 subunits Examples Receptor-ligand interactions Kinase-substrate interactions Transcription factor-co-activator interactions Multiprotein complex, e.g. multimeric enzymes Cramer et al. Science 292:1863, 2001 2 BCHM352, Spring 2011 Significance of protein interaction 3 Most proteins mediate their function through interacting with other proteins To form molecular machines To participate in various regulatory processes Distortions of protein interactions can cause diseases BCHM352, Spring 2011 Yeast two-hybrid Method Pros Bait strain: a protein of interest, bait (B), fused to a DNA-binding domain (DBD) Prey strains: ORFs fused to a transcriptional activation domain (AD) Mate the bait strain to prey strains and plate diploid cells on selective media (e.g. without Histidine) If bait and prey interact in the diploid cell, they reconstitute a transcription factor, which activates a reporter gene whose expression allows the diploid cell to grow on selective media Pick colonies, isolate DNA, and sequence to identify the ORF interacting with the bait High-throughput Can detect transient interactions Cons False positives Non-physiological (done in the yeast nucleus) Can’t detect multiprotein complexes Uetz P. Curr Opin Chem Biol. 6:57, 2002 4 BCHM352, Spring 2011 Tandem affinity purification Method Pros TAP tag: Protein A, Calmodulin binding domain, TEV protease cleavage site Bait protein gene is fused with the DNA sequences encoding TAP tag Tagged bait is expressed in cells and forms native complexes Complexes purified by TAP method Components of each complex are identified through gel separation followed by MS/MS High-throughput Physiological setting Can detect large stable protein complexes Cons High false positives Can’t detect transient interactions Can’t detect interactions not present under the given condition Tagging may disturb complex formation Binary interaction relationship is not clear Chepelev et al. Biotechnol & Biotechnol 22:1, 2008 5 BCHM352, Spring 2011 Large scale protein interaction identification Experimental Yeast two-hybrid Tandem affinity purification Computational Gene fusion Ortholog interaction Phylogenetic profiling Microarray gene co-expression Valencia et al. Curr. Opin. Struct. Biol, 12:368, 2002 6 BCHM352, Spring 2011 Protein interaction data in the public domain Database of Interacting Proteins (DIP) http://dip.doe-mbi.ucla.edu/ The Molecular INTeraction database (MINT) http://mint.bio.uniroma2.it/mint/ The Biomolecular Interaction Network Database (BIND) http://www.binddb.org/ The General Repository for Interaction Datasets (BioGRID) http://www.thebiogrid.org/ Human Protein Reference Database (HPRD) http://www.hprd.org Online Predicted Human Interaction Database (OPHID) http://ophid.utoronto.ca The Munich Information Center for Protein Sequences (MIPS) http://mips.gsf.de 7 BCHM352, Spring 2011 HPRD 8 BCHM352, Spring 2011 Protein interaction networks 9 Saccharomyces cerevisiae Drosophila melanogaster Jeong et al. Nature, 411:41, 2001 Giot et al. Science, 302:1727, 2003 Caenorhabditis elegans Homo sapiens Li et al. Science, 303:540, 2004 Rual et al. Nature, 437:1173, 2005 BCHM352, Spring 2011 Gene regulatory networks Experimental 10 Chromatin immunoprecipitation (ChIP) ChIP-chip ChIP-seq Computational Promoter sequence analysis Reverse engineering from microarray gene expression data Public databases Transfac (http://www.gene-regulation.com) MSigDB (http://www.broadinstitute.org/gsea/msigdb) hPDI (http://bioinfo.wilmer.jhu.edu/PDI/ ) BCHM352, Spring 2011 Shen-orr et al. Nat Genet, 31:64, 2002 KEGG metabolic network 11 BCHM352, Spring 2011 Network visualization tools Cytoscape http://www.cytoscape.org Gehlenborg et al. Nature Methods, 7:S56, 2010 12 BCHM352, Spring 2011 Graph representation of networks Graph: a graph is a set of objects called nodes or vertices connected by links called edges. In mathematics and computer science, a graph is the basic object of study in graph theory. node edge RNA polymerase II 13 Cramer et al. Science 292:1863, 2001 BCHM352, Spring 2011 Undirected graph vs directed graph Protein interaction network Nodes: protein Edges: physical interaction Undirected Krogan et al. Nature 440:637, 2006 Lee et al. Science 298:799, 2002 Metabolic network Transcriptional regulatory network Nodes: metabolites Nodes: transcription factors and genes Edges: enzymes Edges: transcriptional regulation Directed Directed Substrate->Product TF->target gene Ravasz et al. Science 297:1551, 2002 14 BCHM352, Spring 2011 Fhl1 RPL2B Degree, path, shortest path Degree: the number of edges adjacent to a node. A simple measure of the node centrality. Path: a sequence of nodes such that from each of its nodes there is an edge to the next node in the sequence. Shortest path: a path between two nodes such that the sum of the distance of its constituent edges is minimized. YDL176W Degree: 3 Fhl1 Out degree: 4 In degree: 0 15 BCHM352, Spring 2011 Obama vs Lady Gaga: who is more influential? Twitter following (out degree) 16 BCHM352, Spring 2011 Twitter followers (in degree) 701,301 Obama 7,035,548 144,263 Gaga 8,873,525 0 Eminem 3,509,469 Network properties (I): hubs Random network Scale-free network 130 nodes, 215 edges 130 nodes, 215 edges Homogeneous: most nodes have approximately the same number of links Five red nodes with the highest number of links reach 27% of the nodes Heterogeneous: the majority of the nodes have one or two links but a few nodes have a large number of links Five red nodes with the highest degrees reach 60% of the nodes (hubs) Albert et al., Nature, 406:378, 2000 17 BCHM352, Spring 2011 Scale-free biological networks 18 Metabolic network C. elegans Protein interaction network H. sapiens Jeong et al, Nature, 407:651, 2000 Stelzl et al. Cell, 122:957, 2005 BCHM352, Spring 2011 Gene co-expression network S. cerevisiae Noort et al, EMBO Reports,5:280, 2004 Network properties (II): small world network Wichita Stanly Milgram’s small world experiment Social network Average path length between two person Boston Omaha "If you do not know the target person on a personal basis, do not try to contact him directly. Instead, mail this folder to a personal acquaintance who is more likely than you to know the target person." Small world network: a graph in which most nodes can be reached from every other by a small number of steps. Biological interpretation: Efficiency in transfer of biological information Six degrees of separation 19 BCHM352, Spring 2011 Network properties (III): motifs Network motifs: Patterns that occur in the real network significantly more often than in randomized networks. Three-node patterns Milo et al., Science, 298:824, 2002 Feed-forward loop Feedback loop 20 BCHM352, Spring 2011 Network properties (IV): modularity 21 Modularity refers to a group of physically or functionally linked molecules (nodes) that work together to achieve a relatively distinct function. Examples Protein interaction modules Palla et al, Nature, 435:841, 2005 Transcriptional module: a set of coregulated genes sharing a common function Protein complex: assembly of proteins that build up some cellular machinery, commonly spans a dense sub-network of proteins in a protein interaction network Signaling pathway: a chain of interacting proteins propagating a signal in the cell BCHM352, Spring 2011 Gene co-expression modules Shi et al, BMC Syst Biol, 4:74, 2010 Network distance vs functional similarity Proteins that lie closer to one another in a protein interaction network are more likely to have similar function and involve in similar biological process. Sharan et al. Mol Syst Biol, 3:88, 2007 22 BCHM352, Spring 2011 Network-based disease gene prioritization Kohler et al. Am J Hum Genet. 82:949, 2008 For a specific disease, candidate genes can be ranked based on their proximity to known disease genes. 23 BCHM352, Spring 2011 Summary Biological networks Graph representation of networks Graph, node, edge, undirected graph, directed graph, degree, path, shortest path Network properties Hubs and scale-free degree distribution Small-world Motifs Modularity Network-based applications 24 Protein-protein interaction network; Gene regulatory network; Metabolic network Disease gene prioritization BCHM352, Spring 2011