Isolation of N-linked glycopeptides from plasma
advertisement
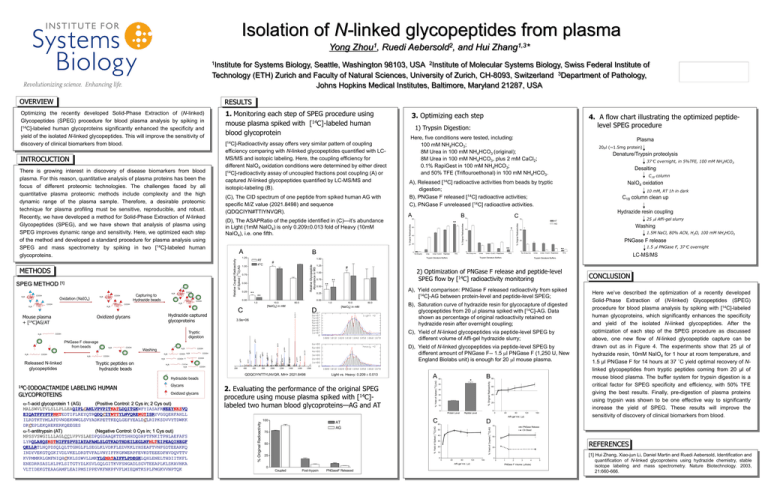
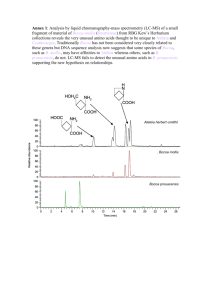
Isolation of N-linked glycopeptides from plasma Yong Zhou1, Ruedi Aebersold2, and Hui Zhang1,3* 1Institute for Systems Biology, Seattle, Washington 98103, USA 2Institute of Molecular Systems Biology, Swiss Federal Institute of Technology (ETH) Zurich and Faculty of Natural Sciences, University of Zurich, CH-8093, Switzerland 3Department of Pathology, Johns Hopkins Medical Institutes, Baltimore, Maryland 21287, USA 1. Monitoring each step of SPEG procedure using mouse plasma spiked with [14C]-labeled human blood glycoprotein PNGase F cleavage from beads H2N COOH Released N-linked glycopeptides H2N H2N COOH Washing H2N Relative Glycopeptide Abundance in MS # H2N COOH H2N COOH H2N Tryptic peptides on hydrazide beads COOH H2N H2N 0.75 0.50 ** 14C-IODOACTAMIDE LABELING HUMAN Oxidized glycans -1-acid glycoprotein 1 (AG) (Positive Control: 2 Cys in; 2 Cys out) MALSWVLTVLSLLPLLEAQIPLCANLVPVPITNATLDQITGKWFYIASAFRNEEYNKSVQ EIQATFFYFTPNKTEDTIFLREYQTRQDQCIYNTTYLNVQRENGTISRYVGGQEHFAHLL ILRDTKTYMLAFDVNDEKNWGLSVYADKPETTKEQLGEFYEALDCLRIPKSDVVYTDWKK DKCEPLEKQHEKERKQEEGES -1-antitrypsin (AT) (Negative Control: 0 Cys in; 1 Cys out) MPSSVSWGILLLAGLCCLVPVSLAEDPQGDAAQKTDTSHHDQDHPTFNKITPNLAEFAFS LYRQLAHQSNSTNIFFSPVSIATAFAMLSLGTKADTHDEILEGLNFNLTEIPEAQIHEGF QELLRTLNQPDSQLQLTTGNGLFLSEGLKLVDKFLEDVKKLYHSEAFTVNFGDTEEAKKQ INDYVEKGTQGKIVDLVKELDRDTVFALVNYIFFKGKWERPFEVKDTEEEDFHVDQVTTV KVPMMKRLGMFNIQHCKKLSSWVLLMKYLGNATAIFFLPDEGKLQHLENELTHDIITKFL ENEDRRSASLHLPKLSITGTYDLKSVLGQLGITKVFSNGADLSGVTEEAPLKLSKAVHKA VLTIDEKGTEAAGAMFLEAIPMSIPPEVKFNKPFVFLMIEQNTKSPLFMGKVVNPTQK C18 column Urea Urea +CaCl2 RapiGest 0 No Enzy me TFE Urea Urea +CaCl2 RapiGest 10 mM, RT 1h in dark C18 column clean up Hydrazide resin coupling 75 25 l Affi-gel slurry AT AG Washing 50 1.5M NaCl, 80% ACN, H2O, 100 mM NH4HCO3 PNGase F release 25 0 No Enzy me TFE NaIO4 oxidation Trypsin Denature Buffers Urea Urea +CaCl2 RapiGest TFE LC-MS/MS Trypsin Denature Buffers 0.00 10.0 50.0 1.0 [NaIO4] in mM C 1.5 l PNGase F, 37C overnight ** A), Yield comparison: PNGase F released radioactivity from spiked [14C]-AG between protein-level and peptide-level SPEG; D 10.0 50.0 [NaIO4] in mM 3.9e+06 B), Saturation curve of hydrazide resin for glycocapture of digested glycopeptides from 20 l plasma spiked with [14C]-AG. Data shown as percentage of original radioactivity retained on hydrazide resin after overnight coupling; C), Yield of N-linked glycopeptides via peptide-level SPEG by different volume of Affi-gel hydrazide slurry; D), Yield of N-linked glycopeptides via peptide-level SPEG by different amount of PNGase F-- 1.5 l PNGase F (1,250 U, New England Biolabs unit) is enough for 20 l mouse plasma. COOH Glycans Desalting COOH QDQCIYNTTYLNVQR, MH+ 2021.8498 Light vs. Heavy: 0.209 0.013 2. Evaluating the performance of the original SPEG procedure using mouse plasma spiked with [14C]labeled two human blood glycoproteins—AG and AT A 60 B * 50 40 30 20 10 80 60 40 20 0 0 Protein Level Peptide Level 0 40 80 120 160 Affi-gel Vol. (l) 100 AT AG 75 50 C 75 50 40 30 20 10 0 40 80 Affi-gel Vol. ( l) Post-trypsin PNGaseF Released PNGase Release On Bead 50 REFERENCES 25 0 0 0 Here we’ve described the optimization of a recently developed Solid-Phase Extraction of (N-linked) Glycopeptides (SPEG) procedure for blood plasma analysis by spiking with [14C]-labeled human glycoproteins, which significantly enhances the specificity and yield of the isolated N-linked glycopeptides. After the optimization of each step of the SPEG procedure as discussed above, one new flow of N-linked glycopeptide capture can be drawn out as in Figure 4. The experiments show that 25 µl of hydrazide resin, 10mM NaIO4 for 1 hour at room temperature, and 1.5 µl PNGase F for 14 hours at 37 ˚C yield optimal recovery of Nlinked glycopeptides from tryptic peptides coming from 20 µl of mouse blood plasma. The buffer system for trypsin digestion is a critical factor for SPEG specificity and efficiency, with 50% TFE giving the best results. Finally, pre-digestion of plasma proteins using trypsin was shown to be one effective way to significantly increase the yield of SPEG. These results will improve the sensitivity of discovery of clinical biomarkers from blood. D 60 25 Coupled CONCLUSION ** COOH COOH H2N 37C overnight, in 5%TFE, 100 mM NH4HCO3 2) Optimization of PNGase F release and peptide-level SPEG flow by [14C] radioactivity monitoring 1.00 COOH Hydrazide beads GLYCOPROTEINS 1.25 Tryptic digestion COOH 20 5 % Original Radioactivity H2N 40 * 10 Trypsin Denature Buffers ** 1.0 Hydrazide captured glycoproteins Oxidized glycans Mouse plasma + [14C]AG/AT Denature/Trypsin proteolysis 14 H2N 60 C 15 % Yield of Coupled [ C]-AG H2N H2N COOH ** No Enzyme 0.25 0.00 COOH B 80 14 COOH 0.50 ** 20l (~1.5mg protein) # 0.75 0.25 A % Yield of Spiked [ C]-AG H2N Oxidation (NaIO4) COOH 1.00 Plasma A), Released [14C] radioactive activities from beads by tryptic digestion; B), PNGase F released [14C] radioactive activities; C), PNGase F unreleased [14C] radioactive activities. 0 1.50 RT 4O C Here, five conditions were tested, including: 100 mM NH4HCO3; 8M Urea in 100 mM NH4HCO3 (original); 8M Urea in 100 mM NH4HCO3, plus 2 mM CaCl2; 0.1% RapiGest in 100 mM NH4HCO3; and 50% TFE (Triflouroethonal) in 100 mM NH4HCO3. ** B 1.25 4. A flow chart illustrating the optimized peptidelevel SPEG procedure 1) Trypsin Digestion: % Yield of Spiked [ 14C]-AG H2N Capturing to Hydrazide beads COOH A % Original Radioactivity COOH [1] H2N (D), The ASAPRatio of the peptide identified in (C)—it’s abundance in Light (1mM NaIO4) is only 0.2090.013 fold of Heavy (10mM NaIO4), i.e. one fifth. 14 METHODS SPEG METHOD (C), The CID spectrum of one peptide from spiked human AG with specific M/Z value (2021.8498) and sequence (QDQCIYN#TTIYNVQR). Relative Coupled Radioactivity There is growing interest in discovery of disease biomarkers from blood plasma. For this reason, quantitative analysis of plasma proteins has been the focus of different proteomic technologies. The challenges faced by all quantitative plasma proteomic methods include complexity and the high dynamic range of the plasma sample. Therefore, a desirable proteomic technique for plasma profiling must be sensitive, reproducible, and robust. Recently, we have developed a method for Solid-Phase Extraction of N-linked Glycopeptides (SPEG), and we have shown that analysis of plasma using SPEG improves dynamic range and sensitivity. Here, we optimized each step of the method and developed a standard procedure for plasma analysis using SPEG and mass spectrometry by spiking in two [14C]-labeled human glycoproteins. of Spiked [ C]-AG INTROCUCTION [14C]-Radioactivity assay offers very similar pattern of coupling efficiency comparing with N-linked glycopeptides quantified with LCMS/MS and isotopic labeling. Here, the coupling efficiency for different NaIO4 oxidation conditions were determined by either direct [14C]-radioactivity assay of uncoupled fractions post coupling (A) or captured N-linked glycopeptides quantified by LC-MS/MS and isotopic-labeling (B). 3. Optimizing each step % Original Radioactivities Optimizing the recently developed Solid-Phase Extraction of (N-linked) Glycopeptides (SPEG) procedure for blood plasma analysis by spiking in [14C]-labeled human glycoproteins significantly enhanced the specificity and yield of the isolated N-linked glycopeptides. This will improve the sensitivity of discovery of clinical biomarkers from blood. % Original Radioactivities RESULTS % Original Radioactivities OVERVIEW 120 160 0 1 2 3 4 PNGase F Volume ( l/tube) 5 [1] Hui Zhang, Xiao-jun Li, Daniel Martin and Ruedi Aebersold, Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nature Biotechnology. 2003, 21:660-666.