RNASeq analysis
advertisement
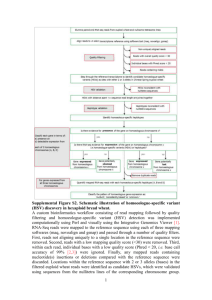
RNAseq analysis Bioinformatics Analysis Team McGill University and Genome Quebec Innovation Center bioinformatics.service@mail.mcgill.ca •Module #: Title of Module •2 Why sequence RNA? • Functional studies – Genome may be constant but experimental conditions have pronounced effects on gene expression • Some molecular features can only be observed at the RNA level – Alternative isoforms, fusion transcripts, RNA editing • Interpreting mutations that do not have an obvious effect on protein sequence – ‘Regulatory’ mutations • Prioritizing protein coding somatic mutations (often heterozygous) Modified from Bionformatics.ca RNA-seq Isolate RNAs Generate cDNA, fragment, size select, add linkers Samples of interest Sequence ends Condition 1 Condition 2 (normal colon) (colon tumor) 100s of millions of paired reads 10s of billions bases of sequence RNA-seq – Applications • Gene expression and differential expression • Transcript discovery • SNV, RNA-editing events, variant validation • Allele specific expression • Gene fusion events detection • Genome annotation and assembly • etc ... RNAseq Challenges • RNAs consist of small exons that may be separated by large introns – Mapping splice-reads to the genome is challenging – Ribosomal and mitochondrial genes are misleading • RNAs come in a wide range of sizes – Small RNAs must be captured separately • RNA is fragile and easily degraded – Low quality material can bias the data Modified from Bionformatics.ca RNA-Seq: Overview RNA-Seq: Input Data Input Data: FASTQ End 1 End 2 Control1_R1.fastq.gz Control2_R1.fastq.gz KnockDown1_R1.fastq.gz Control1_R2.fastq.gz Control2_R2.fastq.gz KnockDown1_R2.fastq.gz KnockDown2_R1.fastq.gz KnockDown2_R2.fastq.gz ~ 10Gb each sample @ERR127302.1 HWI-EAS350_0441:1:1:1055:4898#0/1 GGCTCATCTTGAACTGGGTGGCGACCGTCCCTGGCCCCTTCTTGACACCCAGCG + 4=B@D99BDDDDDDD:DD?B<<=?>6B########################### @ERR127302.2 HWI-EAS350_0441:1:1:1056:1163#0/1 GAATGAGAGGCCCTCCCCGTGGAGGCATGGTATCCGGCCGAGGGGGCTTAGTCA Q = -10 log_10 (p) Where Q is the quality and p is the probability of the base being incorrect. QC of raw sequences QC of raw sequences low qualtity bases can bias subsequent anlaysis (i.e, SNP and SV calling, …) QC of raw sequences Positional Base-Content QC of raw sequences QC of raw sequences Species composition (via BLAST) RNA-Seq: Trimming and Filtering Read Filtering • Clip Illumina adapters: • Trim trailing quality 30 between the adapters and reads. First, Trimmomatic uses a two-step approach to find< matches Trimmomatic a two-step to 16 findbp)matches between thepossible adaptersposition and reads. First, short sections uses of each adapter approach (maximum are tested in each within the short adaptersh k(maximum bp) are i tested in each possible position within the reads.sections h If thi s sof or teach n alignment, own as te16 ‘seed’ a perfect or sufficiently close match, reads. h If thi sby sorthe t n alignment, sh kownparameter as te ‘seed’ i a perfect or sufficiently match, determined seedMismatch (see below), the entire alignmentclose between the determined by theisseedMismatch parameter (see below), entire alignment between the read and adapter scored. This two-step strategy results the in considerable efficiency gains, read is scored.can This results while in considerable efficiencyscore gains, sinceand the adapter seed alignment be two-step calculatedstrategy very quickly, the full alignment is since the seed alignment can be calculated very quickly, while the full alignment score is calculated relatively rarely. calculated relatively rarely. The full alignment score is calculated as follows. Each matching base increases the alignment The is calculated as follows. Each matching increases the alignment scorefull byalignment 0.6, whilescore each mismatch reduces the alignment score bybase Q/10. By considering the • Filter for read length ≥ 32 bp score each mismatch reduces Q/10. By considering the qualitybyof0.6, thewhile base calls, mismatches causedthebyalignment read errorsscore havebyless impact. A perfect match Trimmomatic uses a two-step approach to find matches between the adapters quality thesequence base calls, mismatches read25errors less impact. A perfect of a 12 of base will score just caused over 7, by while baseshave are needed to score 15. As match such and read sections of each 16value bp) aresimple tested inusadellab.org each possible of 12 baseshort sequence will score just7adapter while 25 bases are needed to score 15. As such . position w wearecommend values of between -over 15 as7,(maximum the threshold for alignment mode. RNA-Seq: Mapping Assembly vs. Mapping mapping Reference all vs reference reads contig1 assembly all vs all contig2 RNA-seq: Assembly vs Mapping Reference-based RNA-seq Ref. Genome or Transcriptome RNA-seq reads De novo RNA-seq contig1 contig2 Read Mapping • Mapping problem is challenging: – – – – Need to map millions of short reads to a genome Genome = text with billons of letters Many mapping locations possible NOT exact matching: sequencing errors and biological variants (substitutions, insertions, deletions, splicing) • Clever use of the Burrows-Wheeler Transform http://genomebiology.com/2009/10/3/R25 Genome Biology 2009, Volume 10, Issue 3, Article R25 Langmead et al. R25. increases speed and reduces memory footprint http://genomebiology.com/2009/10/3/R25 Genome Biology 2009, Volume 10, Issue 3, Article R25 •TableOther mappers: BWA, Bowtie, STAR, GEM, etc. Langmead et al. R25.2 1 Bowtie alignment performance versus SOAP and Maq Table 1 Platform CPU time Wall clock time Reads mapped per Bowtie alignment performance versus SOAP and Maq hour (millions) Bowtie -v 2 Platform Server CPU 15 mtime 7s 15 mclock 41 s time Reads 33.8 mapped per Wall SOAP 91 h 57 m 35 s 91 h 47 m 46 s hour 0.10 (millions) Peak virtual memory footprint (megabytes) Bowtie speed-up Reads aligned (%) 1,149virtual memory Peak 13,619 footprint (megabytes) 67.4 aligned (%) Bowtie speed-up Reads 351× 67.3 Bowtie-v 2 Server PC Bowtie Maq SOAP 57s s 1516mm741 s s 1517mm41 9117hh5746mm3535s s 9117hh4753mm467 s 29.5 33.8 0.49 0.10 1,353 1,149 804 13,619 -59.8× 351× 71.9 67.4 74.7 67.3 Bowtie Bowtie 1617mm4158s s 28.8 29.5 1,353 1,353 -- 71.9 71.9 Server PC 1718mm5726s s TopHat: Spliced Reads • Bowtie-based • TopHat: finds/maps to possible splicing junctions. • Important to assemble transcripts later (cufflinks) SAM/BAM Control1.bam Control2.bam SRR013667.1 99 19 8882171 60 76M = 8882214 119 NCCAGCAGCCATAACTGGAAT GGGAAATAAACACTATGTTCAA AG KnockDown1.bam KnockDown2.bam ~ 10Gb each bam SRR013667.1 99 19 8882171 60 76M = 8882214 119 NCCAGCAGCCATAACTGGAATGGG AAATAAACACTATGTTCAAAG • Used to store alignments • SAM = text, BAM = binary Read name Bases Base Qualities Flag Reference Position CIGAR Mate Position SRR013667.1 99 19 8882171 60 76M = 8882214 119 NCCAGCAGCCATAACTGGAATGGGAAATAAACACTATGTTCAAAGCAGA #>A@BABAAAAADDEGCEFDHDEDBCFDBCDBCBDCEACB>AC@CDB@> … The BAM/SAM format samtools.sourceforge.net picard.sourceforge.net Sort,View, Index, Statistics, Etc. $ samtools flagstat C1.bam 110247820 + 0 in total (QC-passed reads + QC-failed reads) 0 + 0 duplicates 110247820 + 0 mapped (100.00%:nan%) 110247820 + 0 paired in sequencing 55137592 + 0 read1 55110228 + 0 read2 93772158 + 0 properly paired (85.06%:nan%) 106460688 + 0 with itself and mate mapped 3787132 + 0 singletons (3.44%:nan%) 1962254 + 0 with mate mapped to a different chr 738766 + 0 with mate mapped to a different chr (mapQ>=5) $ RNA-Seq: Alignment QC RNA-seQc summary statistics broadinstitute.org/cancer/cga/rna-seqc RNA-seQc covergae graph Home-made Rscript: saturation RPKM Saturation Analysis RNA-Seq:Wiggle UCSC: bigWig Track Format • The bigWig format is for display of dense, continuous data that will be displayed in the Genome Browser as a graph. • Count the number of read (coverage at each genomic position: Modified from http://biowhat.ucsd.edu/homer/chipseq/ucsc.html RNA-Seq:Gene-level counts HTseq:Gene-level counts Contro1: 11 reads Control2: 16 reads KnockDown1: 4 reads KnockDown2: 5 reads TSPAN16 • Reads (BAM file) are counted for each gene model (gtf file) using HTSeq-count: www-huber.embl.de/users/anders/HTSeq TSPAN6 TNMD DPM1 SCYL3 C1orf112 FGR CFH FUCA2 … NFYA Control1 11 1 435 203 216 2365 6 380 … 888 Control2 KnockDown1 16 4 0 0 743 836 218 416 643 714 5011 2828 1 4 865 431 … … 827 1674 KnockDown2 5 0 739 352 704 2294 0 523 … 1580 RNA-seq: EDA gqSeqUtils R package: Exploratory Data Analysis 50 MEF_WT_P5 WT 25 MEF_WT_P7 WT ● MEF_WT_P6 WT MEF_KO_P6 KO ● Mutation V2 MEF_KO_P5 KO ● KO 0 WT ● MEF_KO_P7 KO −25 ● MEF_KO_P3 KO −50 MEF_WT_P3 WT −50 −25 0 V1 25 RNA-Seq:Gene-level DGE Home-made Rscript: Gene-level DGE • edgeR and DESeq : Test the effect of exp. variables on gene-level read counts • GLM with negative binomial distribution to account for biological variability (not Poisson!!) DEseq edgeR Differential Gene Expression SYMBOL HNRNPC FAIM2 AC019178 SSC5D GGT5 EXOC3L4 FOXS1 AQP5 SLC27A3 TIMP4 logFC -5.26 -4.82 -6.57 -2.95 -3.03 -3.07 -4.02 -3.73 -2.39 -3.29 PValue 9.19E-55 8.02E-29 2.14E-28 2.39E-27 1.03E-26 9.19E-21 1.69E-19 2.82E-19 6.97E-18 1.21E-17 FDR 5.71E-50 2.49E-24 4.42E-24 3.71E-23 1.28E-22 9.51E-17 1.49E-15 2.18E-15 4.81E-14 7.52E-14 counts.C1 12611 191 100 2274 838 359 113 106 736 126 counts.C2 counts.KD1 counts.KD2 12404 244 443 194 11 3 104 1 1 2123 318 276 803 93 117 344 53 34 92 5 8 113 9 8 637 144 129 120 14 12 Downstream Analyses Pathways/Gene Set (e.g. GOSeq) Regulatory Networks Machine Learning / Classifiers RNA-Seq:Transcriptlevel DGE Cufflinks: transcipt assembly • Assembly: Reports the most parsimonious set of transcripts (transfrags) that explain splicing junctions found by TopHat Cufflinks: transcript abundance • Quantification: Cufflinks implements a linear statistical model to estimate an assignment of abundance to each transcript that explains the observed reads with maximum likelihood. Cufflinks: abundance output • Cufflinks reports abundances as Fragments Per Kilobase of exon model per Million mapped fragments (FPKM) C: Number of read pairs (fragments) from transcript N: Total number of mapped read pairs in library L: number of exonic bases for transcript • Normalizes for transcript length and lib. size Cuffdiff: differential transcript expression • Cudiff – Tests for differential expression of a cufflinks assembly RNA-Seq:Generate report Home-made Rscript Generate report – Noozle-based html report which describe the entire analysis and provide a general set of summary statistics as well as the entire set of results Files generated: – index.html, links to detailed statistics and plots For examples of report generated while using our pipeline please visit our website