Mutational processes molding the genomes of 21 breast cancers
advertisement

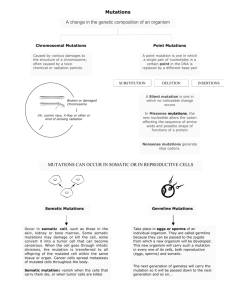
Recent applications of NGS sequencing in cancer studies Andrew Gentles CCSB NGS workshop September 2012 You’ve slogged through QC, trimming, alignment, realignment, variant calling What next ? • Mutational processes molding the genomes of 21 breast cancers/The life history of 21 breast cancers – Nik-Zainal et al. (2012) Cell 149(5):994-1007 • Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia – Jan et al. (2012) Sci Trans Med 4, 149ra118 • Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression – Prensner et al. (2011) Nat Biotech 29: 742-9 Companion papers from Cell May 2012 Whole genome sequencing of 21 Breast cancers Sample PD3851 PD3890 PD3904 PD3905 PD3945 PD4005 PD4006 PD4085 PD4086 PD4088 PD4103 PD4107 PD4109 PD4115 PD4116 PD4120* PD4192 PD4194 PD4198 PD4199 PD4248 Previous Histo patho Age at first histopatholo logical ER Status diagnosis gical Grade diagnosis 61 Ductal III + 41 Ductal III 39 Ductal III + 34 Ductal III 59 Ductal III + 39 Ductal III 39 Ductal III 64 Ductal III + 58 Ductal III 32 Ductal III + 46 Ductal III + 33 Ductal III 67 Ductal III 54 Ductal III + 32 Ductal III + 60 Ductal II + 70 Ductal III 43 Lobular III + 59 Ductal III + 59 Ductal II 48 Ductal II - PR Status HER2 Status + + + + + + + + - + + + + - >30x coverage tumor and normal (188x for *) BRCA mutations BRCA1 BRCA2 BRCA1 BRCA2 BRCA1 BRCA1 BRCA1 BRCA2 BRCA2 Analysis outline • WGS sequencing to >30x coverage tumor/normal – ~100 bp paired-end reads – BWA alignment • Compare tumor/normal for variant calling – CaVEMan, Pindel • Detection of structural rearrangements – In-house method • Inference of copy number changes – ASCAT Summary of somatic mutations • 183916 somatic mutations (SNVs) identified in total • 1372 missense, 117 nonsense, 2 stop-lost, 37 splice, 521 silent • Most frequent mutations in known cancer genes such as TP53, GATA3, PIK3CA, MAP2K4, SMAD4, MLL2, MLL3, NCOR1 Higher rate in BRCA1/2 C>A most common Mutational spectrum in breast cancer Kataegis: regions of enhanced mutation rate Kataegis is highly focal upon zooming in Kataegis associated with structural rearrangements A very deep look into mutation frequencies to reconstruct tumor evolution PD4120a • 188x coverage – enables deep look at mutation frequencies • 70690 somatic substitutions – Some in <5% of reads – Mainly C>* in TpC context – High rate of validation Patterns of copy number alteration in PD4120a Relatively few CNVs Some sub-clonal Mutation frequencies show clusters representing major and minor clones D C B A 1. 35% of reads -> all tumor cells since tumor is 70% tumor (cluster D) 2. Trisomy 1q early since few mutations with high read fraction – most are subclonal 3. 3 major clusters of sub-clonal mutations (A,B,C) 15600 5% 11% 19% 26762 35% Founder clone “most-recent common ancestor” D C B A 4. Cluster C ~19% - more than half of tumor cells (since >1/2*35%) “Pigeonhole principle”: for any 2 mutations, at least one tumor cell must have both – must be on same part of phylogenetic tree If one such mutation in greater fraction than another, must have occurred earlier Cluster C must be on same phylogenetic branch as del13 • If SNVs close enough to SNPs, can be phased with them • 2171 on chr13 • 756 can be phased Phasing of somatic mutations (Supp Fig 4) Phasing of somatic mutations (Supp Fig 4) Found 17 mutually exclusive, 76 examples of sub-clonal evolution Figure 3: Reconstructed evolution of tumor (see paper for details) Sci Trans Med 2012 Prospective separation of residual HSC from leukemic patients Residual HSC lack AML FLT3-ITD mutations Strategy for identifying pre-leukemic mutations in HSC 67-239x exome coverage Occurrence of AML mutations in residual HSC ~25000x targeted coverage Mutations in HSC or both HSC/LSC HSC with the pre-leukemic mutations are capable of differentiating to produce functional immune cells Filtering to identify ncRNAs Enrichment of histone modification marks around transcripts H3K4me2 Figure 2 H3K4me3 Novel transcripts are highly expressed in prostate cancer PCAT-1 is highly expressed in metastatic/high-grade prostate cancer Figure 4b Figure 3f PCAT-1 expression is mutually exclusive with EZH2 Relationship of PCAT-1 to EZH2/PRC complex • RNA-seq discovers novel ncRNAs • PCAT-1 highly expressed in high grade/metastatic prostate cancer • PCAT-1 promotes proliferation • Hypothesized role with EZH2 (c.f. HOTAIR) Final items • Please fill out evaluation form! • Slides: – Available soon from http://ccsb.stanford.edu • Sequence answers forum: – http://seqanswers.com • Stanford discussion group • https://mailman.stanford.edu/mailman/listinfo/wgs_club _stanford