Transposon and Mechanisms of Transposition: Introduction of
advertisement

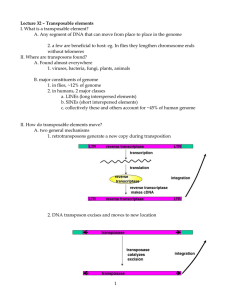
Transposon and Mechanisms of Transposition Transposon DNA sequence that can move in the genome Also called mobile DNA element or transposable element “selfish DNA”--exist only to maintain themselves ? Transposition: The process by which these sequences are copied and inserted into a new site in the genome Probably had a significant influence on evolution How transposon was found 1940s, Barbara McClintock discovered the first transposable element in maize, earned a Nobel prize in 1983. Late 1960s, transposition was also found in Bacteria. Barbara McClintock http://en.wikipedia.org/wiki/Barbara_McClintock Two Categories DNA transposons Retrotransposons “cut-and-paste” “copy-and-paste” Most mobile elements in bacteria is DNA transposons In contrast, most mobile elements in eukaryotes are retrotransposons, but eukaryotic DNA transposons also occur. Lodish et al., Molecular Cell Biology, 7th ed. Fig 10-8 DNA transposons Bacterial Insertion Sequences (IS element) P element in Drosophila General structure of bacterial IS elements Lodish et al., Molecular Cell Biology, 7th ed. Fig 10-9 General process of transposition for DNA transposons Lodish et al., Molecular Cell Biology, 7th ed. Fig 10-10 Retrotransposons LTR retrotransposons: Non-LTR retrotransposons: the most common type of transposons in mammals General structure of eukaryotic LTR retrotransposons Lodish et al., Molecular Cell Biology, 7th ed. Fig 10-11 What is the difference from retrovirus? Generation of RNA from LTR transposon Lodish et al., Molecular Cell Biology, 7th ed. Fig 10-12 Model for reverse transcription Lodish et al., Molecular Cell Biology, 7th ed. Fig 10-13 Retrotransposons Non-LTR retrotransposons long interspersed elements (LINEs) ≈6 kb in human account for 21% of the genome short interspersed elements (SINEs) ≈300 bp in human account for 13% of the genome Lodish et al., Molecular Cell Biology, 7th ed. Fig 10-15 General Principles of LINE transposition Lodish et al., Molecular Cell Biology, 7th ed. Fig 10-16 SINEs (Short Interspersed Elements) Weiner (2000) Fig 1 Most are tRNA derived; Alu is 7SL-RNA Nonautonomous Dependent on other machinery- genome “parasite” RNA Pol III Needs LINE Endonuclease and Reverse Transcriptase for activity Average size 150-200 base pairs Composed of 3 parts 5’ head Body 3’ tail Vassetzky (2013) http://biol.lf1.cuni.cz/ucebnice/images/rep1.gif Kramerov & Vassetzky (2005) Transport Kramerov & Vassetzky (2005) Batzer & Deininger, Nature Reviews Genetics (2002) Box 1 Kramerov & Vassetzky (2005) “Transposons: Mobile DNA” (2012) Where there is a SINE, there is a LINE Specificity of EN/RT of LINE dictates location Expressed during early embryogenesis and decreases in development Active in tumor cells Integrates into germ lines References Batzer, M.A. & Deininger, P.L. Alu repeats and Human genomic diversity. Nature Reviews Genetics 3, 370-379 (2002). Doi:10.1038/nrg798 http://www.nature.com/nrg/journal/v3/n5/box/nrg798_BX1.html Kramerov, D.A. & Vassetzky, N.S. Short Retroposons in Eukaryotic Genomes. International Review of Cytology, vol 247 (2005) doi: 10.1016/S0074-7696/05 Lodish et al., Molecular Cell Biology, 7th ed. “Transposons: Mobile DNA”. (2012) http://users.rcn.com/jkimball.ma.ultranet/BiologyPages/T/Transposons.html Vassetzky. SINEBase (2013) http://sines.eimb.ru Weiner, A. Do all SINEs lead to LINEs? Nature Genetics 24, 332-333 (2000) doi:10.1038/74135 http://www.nature.com/ng/journal/v24/n4/full/ng0400_332.html