Make notes using these questions

Discuss the answers to these questions without referring to your notes.
Then check your notes and see if you can improve your answers.
1.
What is the proteome?
2.
Why is the proteome larger than the genome?
3.
What is ‘alternative RNA splicing’?
4.
How does it affect protein function?
5.
What is ‘post translational modification’?
6.
How does it affect protein function?
AH Biology: Unit 1
Proteomics and Protein
Structure 2
Protein Structure
Think
• What are the functions of protein in a cell?
• Why is protein structure important?
• Why do organisms usually live within a narrow range of temperature and pH?
• If eukaryotic cells are identical in structure relative to their cellular components why do organisms look different?
• How can a gecko stick to Perspex?
• How do multicellular organisms stay together?
Four levels of protein structure
Primary protein structure
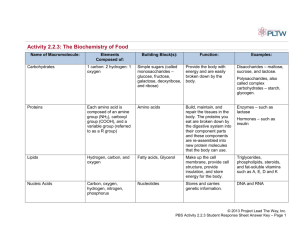
• Proteins are polymers of amino acid monomers. A monomer is the simplest unit of a polymer. There are 20 amino acids in total.
• The primary sequence of a protein is the order in which the amino acids are synthesised by translation into the polypeptide.
Zwitterion: amino acids
• Is a base as the N terminus is free to accept hydrogen ions from solution.
• Is an acid as the C terminus is free to donate hydrogen ions.
• The charge on an amino acid and therefore a protein is pH dependent.
Primary protein structure
• Interactive amino acids
• Peptide bonding
• Primary structure
Peptide bond formation
• A peptide bond is formed between the carboxylic acid (–COOH) terminal of one amino acid and the amine ( –NH
2
) terminal of another.
• This is a condensation reaction as water is produced.
• As a result, all proteins have a carboxyl terminus (end) and an amine terminus.
Secondary protein structure
• Hydrogen bonding along the backbone of the protein strand results in regions of secondary structure called alpha-helices, parallel or antiparallel beta-sheets or turns. These cause the protein to have a threedimensional shape as the linear polypeptide backbone begins to fold.
• Alpha-helices and beta-sheets
Alpha-helix
Hydrogen bonding between the N –H and C=O groups of every 3.5 amino acid residues in the polypeptide backbone.
Beta-sheet
Antiparallel Parallel
Hydrogen bonding between the N –H and C=O groups of the amino acid residues in the polypeptide backbone.
Beta-sheet
Amino acids and R groups
• R groups are the residues or side chains of the 20 amino acids, which have different functional groups.
1.
Positively charged, basic
2.
Negatively charged, acidic
3.
Polar
4.
Hydrophobic, non-polar
5.
Uncharged polar
• These functional groups give the protein its function as they interact with each other and with other structures associated with the protein.
Amino acids and R groups
• R groups
• R group properties test
Tertiary structure
• The polypeptide folds further into a tertiary structure.
• This conformation (shape) is caused by interactions between the R groups.
1.
Hydrophobic interactions
2.
Ionic bonds
3.
Hydrogen bonds
4.
Van der Waals interactions
5.
Disulfide bridges.
• Prosthetic groups (non-protein parts) give proteins added function.
Hydrophobic interactions
• Occur between non-polar R groups along the length of the polypeptide.
• Folding of these regions occurs so that they form a central hydrophobic core , separating non-polar hydrophobic R groups from aqueous solution while the polar hydrophilic R groups are expressed on the outside of the structure, free to interact in aqueous solution.
• Hydrophobic sections of proteins are classically found embedded in the phospholipid bilayer of a cell, while the hydrophilic polar parts are free to interact with the extracellular and intracellular solutions.
Hydrophobic protein domains
Ionic bonds
• Charge dependent attraction occurring between oppositely charged polar R groups, eg between the amino acids arginine and aspartic acid.
• pH affects ionic bonding and results in denaturation of the protein at extremes of pH as the H + and OH
– ions in solution interact with the charge across the ionic bond.
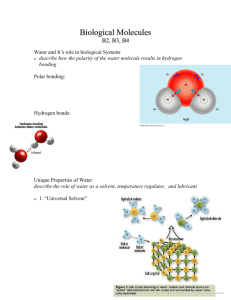
Hydrogen bonds
• Hydrogen bonding is a weak polar interaction that occurs when an electropositive hydrogen atom is shared between two electronegative atoms.
• Hydrogen bonding is charge dependent.
• pH affects hydrogen bonding and results in denaturation of the protein at extremes of pH as the H + and OH
– ions in solution interact with the charge across the hydrogen bond.
Ionic and hydrogen bonds
Van der Waals interactions
• Weak intermolecular force between adjacent atoms.
• Geckos and Van der Waals forces.
Disulfide bridges
• Covalent bonds that form between adjacent cysteine amino acids.
• These can occur within a single polypeptide (tertiary structure) or between adjacent polypeptides (subunits, quaternary structure).
Prosthetic groups
• These are additional non-protein structures that are associated with the protein molecule and give it its final functionality.
• Examples:
1. chlorophyll (magnesium centre), responsible for light capture in photosynthesis
2.
haem (iron centre), found in red blood cells in haemoglobin and responsible for oxygen carriage.
Chlorophyll
Haem
Tertiary structure
• Tertiary structure review
Human pancreatic lipase
Quaternary structure
• Quaternary structure exists in proteins with several connected polypeptide subunits.
• These subunits are held together by all of the interactions listed in the tertiary structure.
• Quaternary structure review
• Immunoglobulins
• Keratin
Haemoglobin: four subunits and four haem groups
Effects of temperature and pH
• Temperature increases the kinetic energy of the protein molecule, placing stress on bonds and breaking them. The weaker intermolecular bonds are particularly susceptible: Van der Waals, hydrogen bonds and ionic bonds.
• Changes in pH affect the concentration of H + and OH – ions in solution. This in turn changes the relative charge of the protein and places stress on polar interactions such as hydrogen bonding and ionic bonding.
• This results in the denaturation of the protein and the loss of tertiary structure and function.
• Denaturation review
Hydrophobic and hydrophilic interactions
• The R groups at the surface of a protein determine its location within a cell.
– Protein trafficking animation of Golgi apparatus.
– Protein transport animation and the enzymes involved.
Pepsin structure: globular protein
Make notes using these questions
1.
Draw the structure of an amino acid molecule. Label the key parts.
2.
Draw a diagram to show how amino acids become joined together. Explain why this is called a ‘condensation’ reaction. What would the opposite reaction by called?
3.
Amino acid R-groups are described as hydrophilic or hydrophobic. What does this mean?
4.
Draw a table to show the four levels of protein structure. Include descriptions and diagrams.
5.
Bonding is extremely important in protein structure. Describe the types of bonds that would be found at each level of protein structure. Which of these are ionic and which are covalent?
6.
How does the type of chemical bonding that occurs in a protein affect its function?
7.
Explain why protein molecules embedded in cell membranes often contain high numbers of hydrophobic R – Groups.
8.
What is a prosthetic group? Give an example.
9.
Not all proteins have a quaternary structure. Explain why.
The fluid mosaic model of membrane structure
The fluid mosaic model of membrane structure
1.
Phospholipid
2.
Cholesterol
3.
Glycolipid
4.
Sugar
5.
Intrinsic transmembrane protein
6.
Intrinsic glycoprotein
7.
Intrinsic protein anchored by a phospholipid
8.
Extrinsic glycoprotein
Mitochondria animation for membrane proteins. Discuss hydrophobic and hydrophillic interactions, and ATP synthase .
The fluid mosaic model of membrane structure
• Regions of hydrophobic R groups allow strong hydrophobic interactions that hold integral proteins, those embedded in the membrane, within the phospholipid bilayer as they are free to interact with the hydrophobic tails of the phospholipids.
• Some integral proteins are transmembrane and cross the phospholipid bilayer, for example:
– channel proteins: facilitated diffusion and active transport
– transporters: sodium potassium pump
– receptors: G-proteins .
The fluid mosaic model of membrane structure
• Peripheral/extrinsic proteins have fewer hydrophobic R groups interacting with the phospholipids.
• Peripheral/extrinsic proteins are responsible for cell–cell interactions:
– cell recognition and the immune system
– junctions such as desmosomes and tight occluding junctions between adjacent cells in tissue are essential for the maintenance of multicellular organisms
– HIV and cell recognition.
HIV and surface proteins
Junctions between cells
Think
• What are the functions of protein in a cell?
• Why is protein structure important?
• Why do organisms usually live within a narrow range of temperature and pH?
• If eukaryotic cells are identical in structure relative to their cellular components why do organisms look different?
• How can a gecko stick to Perspex?
• How do multicellular organisms stay together?