Robert Pirker, MD Genomic Profiling
advertisement

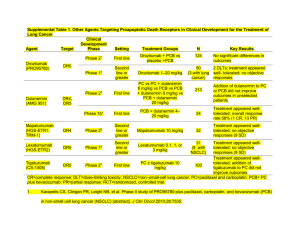
01 Genomic Profiling: Implications for Clinical Research Robert Pirker, MD Traditional Classification and Treatment of NSCLC 02 Classification Treatment: “One-Size-Fits All” Approach Platinum Doublet 1. Li et al. J Clin Oncol. 2013;31(8):1039. 2. NSCLC: ESMO Clinical Recommendations. Ann Oncol. 2007;18:(Supp 2):ii30-ii31. However, NSCLC Is a Group of Heterogeneous and Genetically Complex Tumours Associated with a high rate of somatic mutations 03 Adenocarcinoma Squamous Lawrence et al. Nature. 2013;499:214-18. NSCLC Is Characterised by Various Driver Mutations 04 Li et al. J Clin Oncol. 2013;31(8):1039. There is a Need for a More Precise Treatment Approach Based on Molecular Features in NSCLC 05 Traditional “One-Size-Fits All” Approach All patients with the same diagnosis receive same treatment Precision Medicine Approach Treatment strategy based on patient’s unique genetic profile Genetic Profile A Targeted Therapy Genetic Profile B Standard Therapy • Biologic diversity provides opportunities for exploitation of interpatient tumour heterogeneity by ungrouping a population into molecularly defined subsets in which mutations and/or abnormal gene expressions drive cancer cell growth and survival and can serve as drug targets1 • Treatment decisions made based on tumour phenotype and genomic profile hold the promise to maximize efficacy and minimize toxicity1 Image adapted from http://www.personalizedmedicinecoalition.org/Userfiles/PMC-Corporate/file/pmc_age_of_pmc_factsheet.pdf 1. Li et al. J Clin Oncol. 2013;31(8):1039. Impact of Precision Medicine in the Treatment of Cancer: Personalized vs Non-personalized Therapeutic Strategy Meta-analysis of Phase II single-agent studies (570 trials; 32,149 patients) Schwaederle et al. J Clin Oncol. 2015.61.5997 [Epub ahead of print]. 06 Biomarker-based Precision Medicine Has Significantly Improved Treatment of Patients With NSCLC Targeting cancer drivers improves survival EGFR common mutation+ ALK rearrangement+ 1.Sequist LV et al. J Clin Oncol. 2013;31:3327-3334; 2. Shaw AT et al. N Engl J Med. 2013;368:2385-2394. 07 Consensus Recommendation of EGFR Mutation and ALK Rearrangement Testing in All NSCLC by NCCN, ASCO, and ESMO Lung Treatment Guideline 08 1. NCCN Treatment Guideline for NSCLC v7. 2015. 2. Leighl N et al. J Clin Oncol. 2014;32:3673-3679. 3. Kerr KM et al. Ann Oncol. 2014;25:1681-1690. Types of Biomarkers: Prognostic vs Predictive Biomarkers 09 Prognostic Marker • Information about disease outcome independent of treatment • Example: EGFR mutation in NSCLC • Mutation+ : better prognosis • Mutation- : worse prognosis Predictive Marker • Information on disease outcome related to a specific treatment • Example: EGFR mutation in NSCLC • Mutation+ : ≈70% probability of response to EGFR TKI therapy • Mutation- : <5% probability of response to EGFR TKI therapy Some biomarkers are both prognostic & predictive. Only predictive biomarkers can be used to indicate “which patients should be treated with which drug” (a targeted therapy). Predictive biomarkers can also identify patients who may be harmed by “targeted therapy.” Low and High EGFR Expression in FLEX Pirker R et al. Lancet Oncology 2012, 13, 33 Low HR 0.99 [95% CI 0.84 - 1.16] High HR 0.73 [95% CI 0.58 - 0.93] Interaction p-value=0.044 IPASS: PFS by Mutation Status Mok T et al. NEJM 2009, 361, 947 EGFR Mutation Positive EGFR Mutation Negative HR 0.48 (95% CI o.36-0.64) HR 2.85 (95% CI 2.05-3.98) The Importance of Validation in Biomarker Development 012 • Mutations are abundant in cancer cells • “Driver” mutations are defined as mutations involved in development or progression of a tumour • • Constitute the minority of mutations A subset of drivers (such as EGFR or ALK) and their component cellular pathways may be “actionable” (i.e. have significant diagnostic, prognostic, or therapeutic implications in subsets of cancer patients and for specific therapies • Most mutations do not appear to play a role in cancer progression • “Passenger” mutations are indicative of a high mutation rate resulting from carcinogens and DNA instability Dancey et al. Cell. 2012;148:409. Currently, There Remains a Need for the Identification and Validation of Additional Biomarkers in the Treatment of NSCLC 013 1. Li et al. J Clin Oncol. 2013;31(8):1039. 2. NCCN Clinical Practice Guidelines. NSCLC Version 7.2015 014 Efforts to Identify Actionable Mutations for Treatment of NSCLC Strategies to Identify Biomarkers in NSCLC 015 Oxnard GR et al. J Clin Oncol. 2013;31:1097-1104. 016 Key Technologies Driving the Development of Biomarkers • Advances Beyond 1st-Generation/Sanger Sequencing • Next-Generation Sequencing Advances in Sequencing Technology and Human Genomics 017 Li et al. J Clin Oncol. 2013;31(8):1039. Revolution in Cancer Molecular Diagnosis Involves High-Throughput Technology 018 Li et al. J Clin Oncol. 2013;31(8):1039. Comparison of Traditional (Sanger) and NGS Methods 019 Ross JS et al. Am J Clin Pathol. 2011;136:527-539. The Need for NGS Methods 020 Ross JS et al. Am J Clin Pathol. 2011;136:527-539. 021 Comprehensive Genomic Characterisation and Identification of Targetable Genetic Lesions in NSCLC High Rates of Somatic Mutation Were Seen in Adenocarcinoma of the Lung 022 Somatic mutations (mean 8.9 mutations per megabase) • 230 resected adenocarcinoma of the lung were profiled and analysed as part of the Cancer Genome Atlas initiative Cancer Genome Atlas Research Network. Nature. 2012;511:543-550. New Candidate Driver Mutations Identified in Non-Squamous NSCLC 023 • Aberrations in NF1, MET, ERBB2, and RIT1 occurred in ≈13% of the cases and were enriched in samples lacking an activated oncogene, suggesting a driver role for these events in certain tumours Cancer Genome Atlas Research Network. Nature. 2012;511:543-550. Significantly Mutated Genes in SCC of the Lung 024 • 178 squamous cell carcinomas of the lung were profiled and analysed as part of the Cancer Genome Atlas initiative Cancer Genome Atlas Research Network. Nature. 2012;489:519-525. Potential Targetable Alterations in SCC of the Lung 025 Alterations in targetable oncogenic pathways in SCC of the lung • PI4/RTK/RAS signalling altered in 69% of patients • Alterations in the PI3K/AKT pathway genes were mutually exclusive with EGFR alterations Cancer Genome Atlas Research Network. Nature. 2012;489:519-525. 026 Genomic Profiling in Screening Patients in Biomarker-driven Trials in NSCLC Second-line Therapy in Squamous Lung Cancer (LUNG-MAP): Study Design 027 Fresh tumour biopsy or archival FFPE tumour from eligible patients with stage IIIB or IV lung SCC whose disease has progressed on first-line therapy is evaluated using NGS (FoundationOne) and, in some cases, molecular assays (e.g., IHC-based), carried out in a CLIA-certified laboratory for the presence of drug-specific biomarkers relevant to lung SCC that may serve as targets for drugs currently under study in Lung-MAP. Results are returned within 10–14 days of tissue submission. Herbst RS et al. Clin Cancer Res. 2015;21:1514-1524. Second-line Therapy in Squamous Lung Cancer (LUNG-MAP): Schema for Sub-studies 028 *Archival FFPE tumour, fresh core needle biopsy (CNB) if needed. TT = targeted therapy; CT = chemotherapy (docetaxel or gemcitabine); TKI = tyrosine kinase inhibitor (erlotinib). Herbst RS et al. Clin Cancer Res. 2015;21:1514-1524. ALK Master Protocol in Advanced Lung Adenocarcinoma: Proposed Design 029 A joint NRG/NCI clinical trial to test 2nd-generation ALK inhibitors* PIs: Alice Shaw, MD, Shakun Malik, MD *Ariad: AP26113; Ignyta: RXDX-101; Novartis: ceritinib; Pfizer: PF-0643922; Roche/Chugai: Alectinib; Tesaro: TSR-011; Xcovery: X-396 SWOG Web site. http://swog.org/Visitors/Spring15GpMtg/PlenaryI/02_Abrams.pdf. Accessed August 26, 2015. ALCHEMIST Trial: Trial Design 030 * *NGS to understand the mechanisms of drug resistance and genomic evolution. 1. www.clinicaltrials.gov. Accessed August 26, 2015; 2. Gerber DE et al. Clin Pharmacol Ther. 2015;97:447-650. 031 Genomic Profiling in NSCLC: Key Considerations and Future Perspectives Considerations for Genome Profiling in the Management of Cancer, Including NSCLC 032 • Sampling1 – High intratumour heterogeneity presents challenges1 – Dynamic changes during tumour progression or response to therapy – The need to improve regulatory system to ensure the quality of conducting genetic and genomic testing (quality and quantity of tumour tissues) • Analysis1 – Distinguish between driver mutation from passenger genetic abnormalities – Large computer capacity needed to properly and efficiently analyse the huge amount of data generated • Close collaboration among all stakeholders1 – Multidisciplinary health care providers (surgeons, pulmonologists, radiologists, pathologists, translational scientists, medical oncologists, insurers, regulatory agencies and patients are required for successful clinical implementation of NGS in genotyping and genomic testing1 1. Li T et al. J Clin Oncol. 2013;31:1039-1049. Conclusions 033 • Identification of predictive biomarkers is fundamental for precision medicine to fulfill its promise to maximize efficacy and minimize toxicity – Key driver mutations (i.e. EGFR and ALK) in subpopulations of patients with NSCLC, and their inhibition have resulted in significant improvement in clinical outcomes for such patients • Continued advancement of biomarker-driven precision medicine is challenged by the genetically complexity of tumours and paucity of validated, actionable mutations • Advances in genome technology, especially through NGS, promise rapid and high-throughput ways for large-scale whole genome-wide profiling and analysis with the capacity for better biomarker identification • Its increased use in translational research and clinical development holds the promise of improving our current understanding of oncogenesis and advancing personalised cancer treatment