NMR-assisted computational studies of peptidomimetic inhibitors
advertisement

NMR-assisted computational studies of peptidomimetic inhibitors
bound in the hydrophobic pocket of HIV-1 glycoprotein 41
Miriam Gochin1,2,*, Landon R. Whitby3, Aaron Phillips1 and Dale L. Boger3
Supplementary Data
Index
Figure S1. Structures of the amino acid side chains in the peptidomimetic library
Figure S2. Overlay of different PDB structures of the gp41 hydrophobic pocket
Figure S3. Example docked poses of O2N-{HoPhe]-Nap-OH
Figure S4. AutoDock-Vina docked poses of O2N-{HoPhe]-Ala-OH in receptor 3p7k
Figure S5. AutoDock-Vina docked poses of O2N-{HoPhe]-Ala-OH in receptor 2kp8
Figure S6. AutoDock-Vina docked poses of O2N-{Ala]-Nap-OH in receptor 2r5d
Figure S7. Effect of errors in observed KD , c or protein concentration on PRE and
distance data
Figure S8. Predicted poses of O2N-{HoPhe]-X-OH, X = Phe, Phe4Cl, Nap, Trp, HoPhe
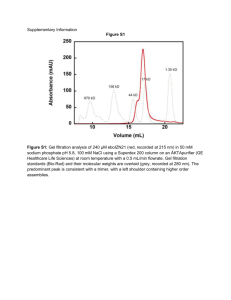
Figure S1
Figure S1. The structures of the 20 natural and unnatural amino acid side chain
substituents used in the synthesis of the 400 member two-subunit α-helix mimetic library.
Figure S2
Figure S2. Overlay of the gp41 hydrophobic pocket region in structures 2r5d (tan), 2r5b
(green), 3p7k (gray) and 2kp8 (pink), created by matching backbone atoms of
hydrophobic pocket residues. Residues in two helices make up the pocket. Sequences
are below, with the hydrophobic pocket region (residues 565-581 as numbered in uniprot
entry P04578) underlined.
2r5d, 2r5b: RMKQIEDKIEEIESKQKKIENEIARIKKLLQLTVWGIKQLQARIL
3p7k:
KIEAIEKKQNNLLRAIEAQQHLLQLTVWGIKQLQARILAVEKKIE
2kp8:
TSLIHSLIEESQNQQEKNEQELLEL-DGDGPQLLSGIVQQQNNLLRAIEAQQHLLQLTVWGIKQLQARILAVE
Figure S3
Figure S3. Examples of low energy poses of O2N-[HoPhe]-Nap-COOH docked into the
gp41 hydrophobic pocket of three receptors, 2r5d (blue), 3p7k (magenta) and 2kp8
(yellow) using Autodock-Vina. Binding scores calculated by Vina are shown in each
panel.
Figure S4
Figure S4. The nine top poses of O2N-[HoPhe]-Ala-OH produced by AutoDock-Vina
docking and scoring in the hydrophobic pocket of 3p7k. The Vina docking score is
indicated in each panel in kcal/mol.
Figure S5
Figure S5. The nine top poses of O2N-[HoPhe]-Ala-OH produced by Vina docking and
scoring in the hydrophobic pocket of 2kp8. The Vina docking score is indicated in each
panel in kcal/mol.
Figure S6.
Figure S6. The nine top poses of O2N-[Ala]-Nap-OH produced by Vina docking and
scoring in the hydrophobic pocket of 2r5d. The Vina docking score is indicated in each
panel in kcal/mol.
Figure S7.
Figure S7. The interplay between c, PRE, fraction of bound ligand, and distance from
spin label. A. Variation of distances calculated from PRE’s as a function of correlation
time used in the calculation, demonstrated for three protons of O2N-[HoPhe]-Ala-OH. B.
RMSD between observed and calculated PRE-derived distances for O2N-[Ala]-Nap-OH
as a function of correlation time for full (green) or half (blue) protein concentration.
Curves through the data points are simply for ease of viewing and have no theoretical
value. The lower RMSD at the expected correlation time of 12ns, based on the size of the
complex, suggests a better fit to the data using the full expected protein concentration. C.
A contour plot showing the range of fb and c over which constant distances could be
obtained from PRE data for O2N-[Ala]-Nap-OH. The protein concentrations used in the
experiment (2, 4, 6, 8, 10, 12uM) were multiplied by factor p to obtain the range of fb’s
shown in the figure. fb = 0.43 at p = ½.
Figure S8
Figure S8. Predicted poses of O2N-[HoPhe]-X-OH shown in black lines, with an
overlaid structure of O2N-[HoPhe]-Ala-OH. A. Receptor 2r5d, X = HoPhe, Trp, Nap; B.
Receptor 3p7k, X = Phe, Phe4Cl, HoPhe, Trp, Nap.