Detection of Single Ring Stage P. falciparum in
advertisement

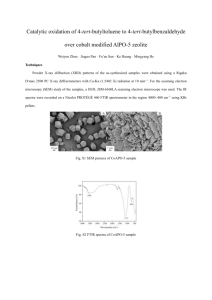
Detection of Single Ring Stage P. falciparum in Human Thin Film Blood Smears Using FTIR Microspectroscopy and Differentiation of Plasmodium Positive from Plasmodium Negative Red Blood Cells by McKale Santin, Dr. Bryan Holmes, Dr. Adam Hunt, and Kenneth A. Puzey of QuantaSpec, Inc. Contact: kpuzey@quantaspec.com ABSTRACT Currently, rapid diagnostic tests for malaria infection perform poorly at low parasite loads, are degraded by severe temperatures, and contain reagents, which contribute to their costs. The overall objective of this study was to perform a preliminary evaluation of the utility of FTIR microspectroscopy for in vitro diagnosis of thin film blood smears for malaria infection. FTIR microspectroscopy has potential advantages in detecting low parasite loads, is not affected by temperature, and does not require any reagents. Giemsa-stained thin film blood smear slides were analyzed in this study. 240 slides with ring stage P. falciparum infected human blood were prepared from culture. P. falciparum negative controls included 80 clinical P. vivax slides (collected and verified by expert microscopy (EM), 40 slides with Salmonella- infected human blood (prepared from culture), and 40 uninfected human blood slides. Infrared spectra were measured from a small area of each slide (~13 microns x 13 microns) usually containing only one red blood cell. Algorithms were written to differentiate Plasmodium positive spectra from Plasmodia negative spectra and tested by cross-validation. The sensitivity was 98.8% to 100% and the specificity was 95.4% to 100% for Plasmodia positive samples with a 95% confidence interval. These results suggest that further study of FTIR spectroscopy as an automated reagent-less diagnostic method with potential for detection of single parasites is warranted. Infrared spectroscopy could radically lower marginal test costs by eliminating the need for expensive consumables. INTRODUCTION Malarial infection is a major global health problem. A key part of malaria control strategies is early case detection with in vitro diagnostics. The current gold standard for malaria diagnosis is expert microscopy of Giemsa-stained blood smears, however this method has many limitations. It is laborintensive, requires consistent, quality staining, and requires diagnosis by a trained, expert microscopist, which are in short supply. Furthermore, most malaria patients are treated in peripheral healthcare facilities that do not have access to quality microscopy. Simple rapid diagnostic tests (RDTs) based on detection of parasite antigens have been introduced to try and provide an alternative to diagnosis with microscopy, yet these diagnostics also have many limitations. RDTs are poor at detecting low-level parasitemia, have a limited shelf life, and contain reagents, which contribute to their cost. Our present study investigates the feasibility of using infrared (IR) microspectroscopy as an alternative diagnostic approach that can overcome the limitations inherent to diagnosis based on analysis of a visible image or by reagent-based assays. Fourier Transform Infrared (FTIR) microspectroscopy can probe the entire chemistry of an intact biological cell with IR light instead of reagents. The spectral signatures of biological cells vary depending on the molecular components of the cell, and the chemical alterations that accompany infection provide the basis for this detection technology. The goal of this research was to evaluate FTIR microspectroscopy for automatically differentiating Plasmodium-positive from Plasmodium-negative red blood cells in thinfilm blood smears. Species/Strain Control Type Parasitemia # of Replicates P. falciparum 7G8 + 6.75% 40 P. falciparum D6 + 5.08% 40 P. falciparum 3D7 + 10.4-12.3% 40 P. falciparum 1776 + 5.8-6.5% 40 P. falciparum HB3 + 7.3-7.7% 40 P. falciparum Dd2 + 5.7% 40 P. vivax, clinical + Variable 40 Salmonella SL1344 - 5 bacteria : 1 RBC 40 Uninfected blood - N/A 40 Table 1. Sample Characterization. Plasmodium-positive controls contained P. falciparum and P. vivax infected human blood. Plasmodiumnegative controls contained Salmonella-infected human blood and uninfected human blood. MATERIALS and METHODS Positive and Negative Controls. Uninfected negative controls: human blood, 5% Hematocrit. Salmonella-infected negative controls: uninfected human blood spiked with Salmonella SL1344. P. falciparum: uninfected blood spiked with strains 7G8, D6 (MR4) and 3D7,1776, HB3, Dd2 (NYU School of Medicine). P. vivax samples were prepared ocular from clinical cases in India Parasite and bacterial counts MCT can be found in Table 1. Sample Preparation. 40 thin detector film blood smears prepared per control group (Table 1). detector 2 aperture All samples prepared on low-e microscope slides (transparent in the visible region but highly reflective in the IR). Samples were fixed and stained with a 10% 74x objective Giemsa solution. Spectral Data Collection. A Bruker IR in sample Hyperion 1000infrared microscope and a Bruker Tensor 27 FTIR spectrometer were used to collect the spectral data (Figures 1 & 2). This system uses a Figure 1. Hyperion 1000 Figure 2. Bruker Hyperion glowbar IR source and a MCT detector. optical beam path. 1000 IR microscope and The microscope was modified with a high power Tensor 27 FTIR spectrometer. reflective objective for an overall magnification of 740X. Spectra collected from a 13x13 m area of a sing-cell layer of each thin film smear (1-3 RBCs). Mid-IR spectra collected from 4000cm-1 to 600cm-1 at a spectral resolution of 2cm-1. 100 scans/measurement. Background measurements taken from an adjacent, blank 13x13 m area of each slide, interactively subtracted from sample spectra using Opus 6.5 software. Data Processing. Raw spectral data organized into two classification groups based on sample identity: Plasmodium-positive and Plasmodium-negative. Data imported into Excel, 1st derivative calculated by taking the slope of the raw data. 1st derivative data imported into JMP software and multivariate discriminant analysis performed on all spectra. Mahalanobis distances calculated for each classification group for each replicate. Algorithm Development. Identification algorithm developed to determine Plasmodium spp. infection based on processed IR spectra. The algorithm consists of a set of vectors that are multiplied with the first derivative spectra of an unknown sample to be identified. For algorithm development, the full set of absorbance values (all optical frequencies) is replaced by a much smaller subset of data containing 350 key optical frequencies for identification. Figure 3. Visual Images, 740X magnification. Ring-stage P. falciparum infected red blood cells (left, red arrow) and uninfected red blood cells (right). Each sample was visually located, then the microscope was switched to IR mode to collect reflectance-absorbance spectra from the center square. Figure 4. Absorbance Spectra. 40 absorbance spectra from P. falciparum strain 3D7 infected blood (left) and 40 absorbance spectra from uninfected human blood (right). RESULTS Visual images of uninfected and ring-stage P. falciparum infected red blood cells at 740X magnification are shown in Figure 3. Spectra were taken from the center square; all other IR light was blocked off by perpendicular apertures. Spectra in the mid-IR region of Plasmodium-positive (P. falciparum strain 3D7) and Plasmodium-negative (uninfected blood) are shown in Figure 4. From the computed Mahalanobis distances, it was found that the longest within-group distances were small (~103) when compared to the shortest across-group distances (~1012). Cross validation testing was used to evaluate the accuracy of the developed algorithm for Plasmodium spp. detection. The algorithm correctly identified 280 out of 280 true positives and 80 out of 80 true negatives. The sensitivity of the developed algorithm was 98.8-100% (95%CI) and the specificity was 95.4-100% (95%CI). CONCLUSION Initial results indicate FTIR microspectroscopy can be used as a rapid identification tool for the detection of Plasmodia in human thin film blood smears with high sensitivity and specificity. All 320 replicates were correctly identified as either malaria positive (240) or malaria negative (80), supporting the hypothesis that FTIR microspectroscopy can be used to detect ring-stage P. falciparum infection. This research study has also demonstrated the potential for infrared microspectroscopy to detect lowlevel parasitemia, as single parasites were detected. We are currently working on collecting a larger clinical sample set, and increasing the number of red blood cells that can be diagnosed simultaneously. ACKNOWLEDGEMENTS This work is supported by the U.S Army Medical Research and Materiel Command under contract No.W81XWH09-C-0019. The views, opinions and/or findings contained in this report are those of the authors and should not be construed as an official Department of the Army position, policy or decision unless so designated by other documentation. “In the conduct of research where humans are the subjects, the investigator(s) adhered to the policies regarding the protection of human subjects as prescribed by Code of Federal Regulations (CFR) Title 45, Volume 1, Part 46; Title 32, Chapter 1, Part 219; and Title 21, Chapter 1, Part 50 (Protection of Human Subjects).” Automated Reagent-less Differentiation of P. falciparum from P. vivax in Human Thin Film Blood Smears With FTIR Microspectroscopy by Kenneth A. Puzey, Dr. Bryan Holmes, Dr. Adam Hunt, and McKale Santin of QuantaSpec, Inc. Contact: kpuzey@quantaspec.com ABSTRACT MATERIALS and METHODS Positive and Negative Controls. Uninfected negative controls: human blood, 5% Hematocrit. Salmonella-infected negative controls: uninfected blood spiked with Salmonella SL1344. P. falciparum: uninfected blood spiked with strains 7G8, D6 (MR4) and 3D7,1776, HB3, Dd2 (NYU School of Medicine). P. vivax samples were prepared from clinical cases in India. Sample Preparation. 40 thinfilm blood smears prepared per control group. All samples prepared on low-e microscope slides (transparent in the visible region but highly reflective in the IR). Samples were fixed and stained with a 10% Giemsa solution. Spectral Data Collection. A Bruker Hyperion 1000infrared microscope and a Bruker Tensor 27 FTIR spectrometer were used to collect the spectral data (Figures 1 & 2). This system uses a glowbar IR source and a liquid nitrogen-cooled MCT detector. The microscope was modified with a high power reflective objective for an overall magnification of 740X. Spectra collected from a 13x13 m area of a sing-cell layer of each thin film smear (1-3 RBCs). Mid-IR spectra collected from 4000cm-1 to 600cm-1 at a spectral resolution of 2cm-1. 100 scans/measurement. Background measurements taken from an adjacent, blank 13x13 m area of each slide, interactively subtracted from sample spectra using Opus 6.5 software. Data Processing. Raw spectral data organized into 3 classification groups based on the identity of the spectral sample: P. falciparum-positive, P. vivaxpositive, and Plasmodia-negative. Data imported into Excel, 1st derivative calculated by taking the slope of the raw data. 1st derivative data imported into JMP software and multivariate discriminant analysis performed on all spectra. Mahalanobis distances calculated for each classification group for each replicate. Algorithm Development. Identification algorithms developed to determine P. INTRODUCTION falciparum infection, P. vivax infection, or no infection based on processed IR spectra. The algorithms Over 3 billion people worldwide are at risk of malaria, representing almost half of the world’s consists of a set of vectors that are multiplied with the first derivative spectra of an unknown sample to population. Prompt and correct diagnosis of malarial infection is a primary part of malaria control and is be identified. For algorithm development, the full set of absorbance values (all optical frequencies) is essential for saving patient lives. In regions where both Plasmodium falciparum and Plasmodium replaced by a much smaller subset of data containing 350 key optical vivax are present, effective diagnosis requires not only detecting malaria infection but also determining frequencies for identification. the species of infection, as different species respond to different chemotherapeutic treatments. Expert microscopy remains the gold standard for distinguishing different species of malarial infection, but Figure 3. Visual Images, 740X unfortunately high-quality expert microscopy is difficult to maintain in resource-poor settings where the magnification. P. vivax infected majority of malaria diagnosis is being performed. Rapid diagnostic tests (RDTs) based on detection of red blood cells (left, blue arrow) species-specific antigens such as pLDH (parasite lactate dehydrogenase) have been introduced to and P. falciparum infected red provide an alternative to diagnosis with microscopy. However, RDTs have many limitations. They are blood cells (right, red arrow). poor at detecting low-level parasitemia, have a limited shelf life, and contain reagents, which contribute Each sample was visually located, to their cost. Our present study investigates the feasibility of using infrared (IR) microspectroscopy as then the microscope was switched an alternative diagnostic approach that can overcome the limitations inherent to diagnosis based on to IR mode to collect reflectanceanalysis of a visible image or by reagent-based assays. Fourier Transform Infrared (FTIR) absorbance spectra from the microspectroscopy can probe the entire chemistry of an intact biological cell with IR light instead of center 13x13 m square. reagents. The spectral signatures of biological cells vary depending on the molecular components of the cell, and the chemical alterations that accompany infection provide the basis for this detection RESULTS technology. The goal of this research was to evaluate FTIR microspectroscopy for automatically Visual images of P. vivax and P. falciparum infected red blood cells at 740X magnification are differentiating Plasmodium falciparum from Plasmodium vivax infected red blood cells shown in Figure 3. Spectra were taken from the center square; all other IR light was blocked off by in thin- film human blood smears. perpendicular apertures. From the FTIR absorbance spectra, P. vivax and P. falciparum cannot be visually distinguished. From multivariate analysis, Mahalanobis distances were calculated for each replicate to every other replicate. It was found that the longest within-group distances are small (~103) Ocular Camera when compared to the shortest across-group distances (~1012). Cross validation testing was used to evaluate the accuracy of the developed algorithm for Plasmodium spp. detection. The algorithm correctly identified 240 out of 240 true positives and 80 out of 80 true negatives. The sensitivity of the P.f. identification algorithm was 98.4-100% (95%CI) and the specificity was 97.7-100% MCT detector (95%CI). The sensitivity of the P.v. identification algorithm was 95.4%-100% (95%CI) and the specificity was 98.8%-100%(95%CI). In malaria cases species of infection affects course of treatment. Differentiation of P. falciparum from P. vivax by RDTs requires multiple antibodies, which increases test costs. Furthermore, RDTs are subject to reader error. Speciation by visual microscopy is dependent on the skill and availability of an expert microscopist. The objective of this study was to evaluate the utility of FTIR microspectroscopy for automatic reagent-less differentiation of P. falciparum from P. vivax infected human red blood cells. Geimsa-stained thin film blood smear slides were analyzed in this study. For P. falciparum positive controls, 240 slides with ring stage P. falciparum were prepared from culture. For P. vivax positive controls, 40 clinical P. vivax slides were collected and verified by expert microscopy (EM). For negative controls, 40 slides with Salmonella-infected blood (prepared from culture) and 40 uninfected blood slides were prepared. Infrared spectra were measured from a small area of each slide (~13 microns x13 microns) typically containing only one red blood cell. Algorithms were written to differentiate red blood cells infected with P. falciparum, red blood cells infected with P. vivax, red blood cells infected with Salmonella and uninfected red blood cells based on their infrared spectra. Algorithms were tested by cross-validation. For P. falciparum sensitivity was 98.4 to 100% and specificity was 97.7% to 100% (95% CI). For P. vivax the sensitivity was 95.4% to 100% and the specificity was 98.8% to 100% (95% CI). These results suggest that FTIR spectroscopy may be useful for automated reagent-less differentiation of malaria infection. In high throughput settings spectroscopy testing may be lower cost because it does not require consumables. CONCLUSION aperture Detector 2 Initial results indicate that RBCs infected with P.f. can be differentiated from RBCs infected with P.v. in thin film blood smears using FTIR microspectroscopy with high sensitivity and specificity. This method is reagent-less and automated (results provided by computer) and is capable of detecting a single Plasmodia parasite. Further study with slides from both clinical P.f. and clinical P.v. from a larger number of cases will be needed to determine the clinical utility of FTIR microspectroscopy for diagnosis and such a study is underway. Equipment modifications to examine a large number of RBCs in parallel are also underway to improve diagnostic throughput. 74x Objective Sample IR in Figure 1. Hyperion 1000™ optical beam path. ACKNOWLEDGEMENTS Figure 2. Bruker Hyperion 1000™ IR microscope and Tensor™ 27 FTIR spectrometer. This work is supported by the U.S Army Medical Research and Materiel Command under contract No.W81XWH09-C-0019. The views, opinions and/or findings contained in this report are those of the authors and should not be construed as an official Department of the Army position, policy or decision unless so designated by other documentatio “In the conduct of research where humans are the subjects, the investigator(s) adhered to the policies regarding the protection of human subjects as prescribed by Code of Federal Regulations (CFR) Title 45, Volume 1, Part 46; Title 32, Chapter 1, Part 219; and Title 21, Chapter 1, Part 50 (Protection of Human Subjects).” Automated Reagent-less Differentiation of Three Drug Susceptible Strains of P. falciparum from Three Drug Resistant Strains of P. falciparum in Human Thin Film Blood Smears Using FTIR Microspectroscopy by Kenneth A. Puzey, Dr. Bryan Holmes, Dr. Adam Hunt, and McKale Santin of QuantaSpec, Inc. Contact: kpuzey@quantaspec.com ABSTRACT In some regions of the world malaria parasite drug resistance is present in 50% of cases. Unfortunately, tests to determine drug resistance are not clinically available forcing health ministries and doctors to make difficult choices. An economical clinical test for drug resistance would enable doctors to administer less expensive chloroquine to susceptible cases, lowering health costs and slowing the spread of resistance to newer drugs. The objective of this study was a preliminary evaluation of the utility of FTIR microspectroscopy for differentiating red blood cells infected with drug resistant strains and drug susceptible strains of P. falciparum. 120 Geimsa-stained thin film blood smear slides were prepared with drug-susceptible ring stage P. falciparum from culture (40 slides strain 3D7, 40 slides strain 1776, 40 slides D6), and 120 Geimsa-stained thin film blood smear slides were prepare with drug-resistant ring stage P. falciparum from culture (40 slides strain HB3, 40 slides strain Dd2, 40 slides strain 7G8). Negative controls included 40 Geimsa-stained thin film blood smear slides of uninfected human blood as well as human blood infected with Salmonella from culture (40 slides). Additional P. falciparum negative controls included 40 clinical Geimsa-stained P. vivax slides collected and verified by expert microscopy (EM). Infrared spectra were measured from a small area of each slide (~13 microns x13 microns) typically containing only one red blood cell. Algorithms were written to differentiate red blood cells infected with P. falciparum, red blood cells infected with P. vivax, red blood cells infected with Salmonella and uninfected red blood cells based on their infrared spectrum. Algorithms were tested by cross-validation. For drug susceptible strains, sensitivity was 97% to 100% and specificity was 98.7% to 100% (95% CI). For drug resistant strains sensitivity was 97% to 100% and specificity was 98.7% to 100% (95% CI). These results suggest that FTIR spectroscopy may be useful for automated reagent-less differentiation of drug resistant and drug susceptible strains of P. falciparum in thin film blood smears. This capability could enable more cost effective case management and reduce the spread of drug resistance to newer drugs. microscope slides (transparent in the visible region but highly reflective in the IR). Samples were fixed and stained with a 10% Giemsa solution. Spectral Data Collection. A Bruker Hyperion 1000infrared microscope and a Bruker Tensor 27 FTIR spectrometer were used to collect the spectral data (glowbar IR source/ liquid nitrogen-cooled MCT detector). 74X reflective objective used to collect spectra from a 13x13 m area of a sing-cell layer of each thin film smear (1-3 RBCs). Mid-IR spectra collected from 4000cm-1 to 600cm-1 at a spectral resolution of 2cm-1. 100 scans/measurement. Background measurements taken from an adjacent, blank 13x13 m area of each slide, subtracted using Opus 6.5 software. Data Processing. Raw spectral data organized into 3 classification groups based on the identity of the spectral sample: drug susceptible P.f.-positive, drug-resistant P.f.-positive, and P.f.-negative. Data imported into Excel, 1st derivative calculated by taking the slope of the raw data. 1st derivative data imported into JMP software and multivariate discriminant analysis performed on all spectra. Mahalanobis distances calculated for each classification group for each replicate. Algorithm Development. Identification algorithms developed to differentiate drug-susceptible P.f.positive from drug- resistant P.f.- positive from P.f.-negative infection based on processed IR spectra. The algorithm consists of a set of vectors that are multiplied with the first derivative spectra of an unknown sample to be identified. For algorithm development, the full set of absorbance values (all optical frequencies) is replaced by a much smaller subset of data containing 350 key optical frequencies for identification. Figure 1. Absorbance Spectra. 40 absorbance spectra from drug susceptible P. falciparum D6 (left) and 40 absorbance spectra from drug resistant P. falciparum Hb3 (right). INTRODUCTION Each year, there are an estimated 250 million malaria cases and approximately 1 million malariarelated deaths. Fundamental to reducing the burden of malaria infection and improving patient outcome is rapid and accurate diagnosis. Field diagnosis and treatment of malarial infection in malaria-endemic regions remains a problem, and is becoming increasingly difficult due to malaria parasite drug resistance. Major methods for malaria diagnosis (expert microscopy and rapid diagnostic tests) are unable to detect drug resistance prior to treatment, and instead are used to monitor for treatment failure. This method is time consuming, with prolonged periods of patient followup, and is costly due to the need for multiple tests. Regardless of the species of malaria, all drug resistance mechanisms involve genetic and chemical differences in the parasite. Our present study investigates the feasibility of using infrared (IR) microspectroscopy as an alternative diagnostic approach that can overcome the limitations inherent to diagnosis based on analysis of a visible image or by reagent-based assays. Fourier Transform Infrared (FTIR) microspectroscopy can probe the entire chemistry of an intact biological cell with IR light instead of reagents. The chemical differences between strains provides the basis for this detection technology. The goal of this research was to evaluate FTIR microspectroscopy for automatically differentiating drug susceptible P. falciparum from drug resistant P. falciparum in thin-film blood smears. Mahalanobis Distances Actual 3D7 7G8 1776 D6 Dd2 HB3 P. vivax Salmonella UIB 3D7 1.8e4 7.8e12 3.5e13 1.9e13 5.7e13 3.8e13 2.3e14 3.1e13 1.8e13 7G8 7.8e12 5.5e3 1.1e13 55e12 2.6e13 1.3e13 1.6e14 1.1e13 3.8e12 1776 3.5e13 1.1e13 1.1e4 4.7e12 3.4e12 2.8e11 8.7e13 7.4e11 3.7e12 D6 1.9e13 5.5e12 4.7e12 1.6e4 1.2e13 6.3e12 1.2e14 2.9e12 1.6e12 Dd2 5.7e13 2.6e13 3.4e12 1.2e13 7.3e3 2.5e12 6.0e13 3.9e12 1.2e13 HB3 3.8e13 1.3e13 2.8e11 6.3e12 2.5e12 3.4e3 8.2e13 1.1e12 4.9e12 P. vivax 2.3e14 1.6e14 8.7e13 1.2e14 6.0e13 8.2e13 7.2e3 9.3e13 1.2e14 Salmonella 3.1e13 1.1e13 7.4e11 2.9e12 3.9e12 1.1e12 9.3e13 1.2e4 2.5e12 UIB 1.8E13 3.8e12 3.7e12 1.6e12 1.2e13 4.9e12 1.2e14 2.5e12 2.4e4 Figure 2. 1st Derivative of Absorbance Spectra. 40 1st derivatives from drug susceptible P. falciparum D6 spectra (left) and 40 1st derivatives from drug resistant P. falciparum Hb3 spectra (right). RESULTS Spectra in the mid-IR region of drug susceptible P. falciparum D6 and drug resistant P. falciparum Hb3 are shown in Figure 1. From the FTIR absorbance spectra, drug susceptible and drug resistant species cannot be visually distinguished. Figure 2 shows the 1st derivative of the absorbance data for drug susceptible P.f D6 and drug resistant P.f Hb3. Table 1 shows the computed Mahalanobis distances using all optical frequencies. The chart shows within-group maximum distances (green, diagonal values) and between-group minimum distances (red values). It was found that the longest within-group distances are small (~103) when compared to the across-group shortest distances (~1012). Cross validation testing was used to evaluate the accuracy of the developed algorithms for drug susceptible P.f positive, drug resistant P.f positive, and P.f negative. The algorithm correctly identified 120/120 drug susceptible P.f replicates, 120/120 drug resistant P.f replicates and 120/120 P.f negatives. The sensitivity of the developed algorithm for drug-susceptible P.f. was 97-100% (95%CI) and the specificity was 98.7-100% (95%CI). The sensitivity of the developed algorithm for drug-resistant P.f. was 97-100%(95%CI) and the specificity was 98.7-100% (95%CI). CONCLUSION Table 1. Calculated Mahalanobis Distances. The vertical column represents the actual identity of the sample and the horizontal row represents the comparison group. Values were calculated using all optical frequencies measured from 4000-600cm-1. Values in green represent the furthest distance between samples in the same identity group, and values in red represent the closest distance between sample from different identity groups. Initial results indicate that FTIR IR microscopy can differentiate drug-susceptible and drug-resistant strains of P.f. (for the strains that were studied). Additional studies with additional strains would be of interest to determine if the initial results are valid for a wider variety of strains. In addition, it was found that IR spectra could be used to accurately differentiate all six strains from each other as well as from P.v. and negative controls with sensitivity of 91.19-100%(95%CI) and with a specificity of 98.98100%(95%CI). Therefore, further study of FTIR microspectroscopy as an alternative diagnostic method is warranted. MATERIALS and METHODS ACKNOWLEDGEMENTS Positive and Negative Controls. Uninfected negative controls: human blood. Salmonella/ P. vivax infected negative controls: uninfected blood spiked with Salmonella SL1344, P. vivax clinical cases from India. P. falciparum drug susceptible: uninfected blood spiked with strains 3D7, or 1776, or D6. P. falciparum drug resistant: uninfected blood spiked with strains HB3, or Dd2, or 7G8. Sample Preparation. 40 thin-film Blood smears prepared per group. All samples prepared on low-e This work is supported by the U.S Army Medical Research and Materiel Command under contract No.W81XWH09-C-0019. The views, opinions and/or findings contained in this report are those of the authors and should not be construed as an official Department of the Army position, policy or decision unless so designated by other documentation. “In the conduct of research where humans are the subjects, the investigator(s) adhered to the policies regarding the protection of human subjects as prescribed by Code of Federal Regulations (CFR) Title 45, Volume 1, Part 46; Title 32, Chapter 1, Part 219; and Title 21, Chapter 1, Part 50 (Protection of Human Subjects).”