Document
advertisement

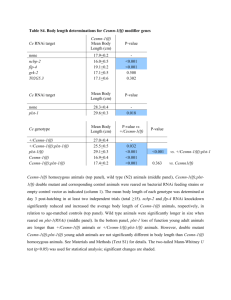
BEH.109: Laboratory Fundamentals in Biological Engineering. MODULE 3 Eukaryotic Cells as Phenotypic Indicators: The use of RNAi to modulate gene expression Instructor: Leona D. Samson Teaching Assistants: Jenn Cheng and Lisa Joslin With additional invaluable help from Lisa Smeester and Rebecca Fry Monday March 31 DAY 1 Tues April 1 DAY 1 Wed April 2 DAY 2 Thurs April 3 DAY 2 Module 3 Overview & mini-lecture on RNAi Module 3 Overview & mini-lecture on RNAi Comprehensive lecture on RNAi with some examples Comprehensive lecture on RNAi with some examples Safety Orientation Safety Orientation Sterile Technique Sterile Technique Transfection of EGFP & p53 siRNA into EGFP expressing HeLa cells Transfection of EGFP & p53 siRNA into EGFP expressing HeLa cells Harvest transfected cells Microscope analysis & FACS analysis Analyze data Harvest transfected cells Microscope analysis & FACS analysis Analyze data Monday April 7 DAY 3 Tues April 8 DAY 3 Wed April 9 DAY 4 Thurs April 10 DAY 4 Introduction to the ATM, ATR, EXO1 and AAG genes Introduction to the ATM, ATR, EXO1 and AAG genes Ambion and Blast session to design new siRNAs for four genes. Ambion and Blast session to design siRNAs for four genes. siRNA is ordered for next experiment siRNA is ordered for next experiment Monday April 14 DAY 5 Tues April 15 DAY 5 Label isolated RNA and hybridize to microarray slides Label isolated RNA and hybridize to microarray slides Introduction to DNA microarrays and overview of what will happen on days 5 & 6 Introduction to DNA microarrays and overview of what will happen on days 5 & 6 Transfect four new si.RNAs; cellular RNA will be isolated over the w/e Transfect four new si.RNAs; cellular RNA will be isolated over the w/e Informal Presentation of FACS data by students Wed April 16 DAY 6 Informal Presentation of FACS data by students Thurs April 17 DAY 6 Scan microarray slides and analyze results Scan microarray slides and analyze results Wed April 23 DAY 7 Patriots Day MIT Holiday MODULE 3 Student Presentations Thurs April 24 DAY 7 MODULE 3 Student Presentations Snapshot of the next four weeks We will eliminate the expression of six different genes using RNAi technology, human cells, fluorescent proteins and DNA microarrays You WILL be required to write a lab report, but Class still happens on April 24rd. Please follow the "BEH.109 Guidelines for Module Reports" handout that was given to you previously. The report will be DUE ON MONDAY, APRIL 29th BY 5 PM. RNA Interference - RNAi BE109 Module 3 Day 2 lecture RNA interference first discovered in Petunias! Called PTGS, for “Post Transcriptional Gene Silencing” Color changes can be induced by RNAi, or PTGS.. Post transciptional gene silencing Small (21-23 nts) RNA duplexes, with the same sequence as in the silenced gene, were identified as being responsible for knocking down expression So what other organisms can do this thing called PTGS? “Post Transcriptional Gene Silencing” Arabidopsis thaliana Protozoa can use RNAi for gene silencing Planaria Trypanosomes Hydra RNAi can operate in insects too! Sulston and Horvitz (1977). Develop. Biol. 56, 110-156. RNAi is used by C. elegans to control the timing of development of various tissues Such gene silencing is a natural phenomenon in this organism 19 nt duplex 2 nt 3’ overhangs This dsRNA species found in plants, C. elegans and Drosophila melanogaster undergoing gene silencing….but how to prove it is responsible? Purified them and showed in vitro silencing in Drosophila extracts; used sythetic sdRNA oligo to achieve same thing! C. Elegans movie C. Elegans grow on agar dishes with E. coli bacteria as a source of food. If they eat E. coli expressing dsRNA molecules…this creates specific knock-down of gene expression! In C. elegans the siRNA effects can be amplified making the silencing quite stable This does not appear to happen in mammalian cells (RISC = RNAi Induced Silencing Complex; RdRP = RNA dependent RNA polymerase) http://www.nature.com/cgi-taf/DynaPage.taf?file=/nature/journal/v421/n6920/full/421220a_fs.html&content_filetype=pdf 19,757 genes 16,757 have been inactivated by RNAi 10% display spontaneous phenotype; this 10% is enriched for conserved genes 19,757 genes 16,757 knock down mutants were screened for body fat content 305 knock downs had increased body fat, 112 genes had decreased..new targets for obesity? C. Elegans movie So…what about RNAi in mammalian cells… May 2001…the first report… How does RNAi work in mammalian cells? RNAi works postranscriptionally…….. in key two steps! Model for RNAi siRNA step one: processing the dsRNA into 21-23 nt fragments QuickT ime ™an d a GIF deco mpre ssor ar e need ed to see this pictur e. 34 27 21 20 16 short-interfering RNA Tuschl, 2001 Dicer contains two RNAse III domains long dsRNA siRNAs siRNAs have a defined structure 19 nt duplex 2 nt 3’ overhangs step two: the antisense strand of the siRNA guides cleavage Tuschl, 2002 RNAi silencing complex • may be associated with translating ribosomes • active RNAse enzyme not yet identified • may participate in endogenous pathways that silence genes via translational repression Mammals exhibit potent responses to dsRNA P P dsRNA PKR interferon production P eiF2a apoptosis Blockage of protein synthesis cell death smaller RNAs can escape the PKR pathway recall that siRNAs are intermediate effectors In the RNAi pathway siRNAs are not recognized by the PKR! Two ways to get double stranded RNA lentiviral construct for siRNAs Rubinson et al Nature Genetics, 2003 Practical Aspects of RNAi • biological research – defining gene function (gene knockout) • C. elegans genome RNAi projects – defining biochemical pathways • microarray screening of RNAi knockouts • therapeutic treatment – cancer – viral infection – parasitic infection HIV levels can be reduced by 30-50 fold by siRNA!!! What is the killer virus that might sweep the globe? Will RNAi eventually become an effective and specific antiviral therapy?