One method to study a gene`s function is to knock out the
advertisement

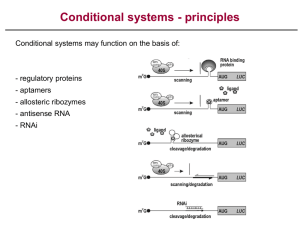
Using RNA Interference to Knock-Down Lis1, Gl, and Ard1 in Drosophila Tramy Truong Mentor: Rahul Warrior One method to study a gene’s function is to knock out the expression of the gene and observe the phenotype. A common technique to reduce the wild-type expression level of a specific gene is the use of the RNA interference system. RNA interference, or RNAi, is a system in which small interfering RNA molecules (siRNAs) function to inactivate gene expression by degrading mRNAs of the gene of interest and, thereby, blocking translation. However, not all genes are susceptible to RNAi silencing, and confidence in this system remains low among researchers. This experiment tested the efficiency of the RNAi system in Drosophila using genes with known functions: Lis1, Gl, and Ard1. The GAL4/UAS system was used to express the RNAi in a tissue-specific manner. By comparing observed phenotypes to known phenotypes, the RNAi system was revealed to be effective at down regulating Lis1, Gl, and Ard1 genes in the germline. Validating RNAi efficiency will not only provide scientists with a certainty that the system works, but will also allow them to apply the system in other tissues with unknown phenotype to learn about the gene’s role in that specific tissue.