RNAinterference (RNAi) is an evolutionary conserved mechanism
advertisement

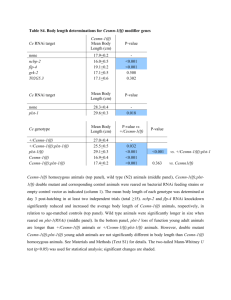
RNA interference: basic concepts and application to ion channel Annarosa Arcangeli Dip. Patologia e Oncologia Sperimentali Università di Firenze RNA interference (RNAi) is an evolutionary conserved mechanism that uses short antisense RNAs that are generated by “dicing” dsRNA precursors to target the corresponding mRNAs for cleavage. First discovered in plants, where it was known as post-transcriptional gene silencing, RNAi occurs in a wide variety of eukaryotic organisms, such as the nematode Caenorabditis elegans and Drosophila. It was the elucidation of the mechanisms underlying RNAi in such species that led to the realisation of its potential as a power research tool for manipulating gene expression1,2. In fact, RNAi is becoming nowadays one of the most powerful tool to determine gene function as well as to knock down undesired gene transcripts in Gene Therapy protocols. Different methods of expressing the short double-stranded (ds) RNA intermediates required for interference in mammalian systems have been developed, including the introduction of short interfering (si)RMAs by direct transfection or driven from transfected plasmids or lentiviral vectors encoding short hairpin (sh)RNAs. Although RNAi relies upon a high degree of specificity, recent findings suggest that offtarget non-specific effects can be encountered. We tested the effect of RNAi on potassium channels of the hERG1 family; such channels are overexpressed in a high variety of human cancers, and their activity regulates tumour cell proliferation and invasiveness 3. Two splicing variants of the hERG1 gene have been discovered so far, the hERG1a and the hERG1b variant. The latter is a N-terminus deleted isoform, that is specifically expressed in some types of human tumours, like leukemias and neuroblastomas 4. We tested 4 synthetic siRNAs, mapping in the N-terminus domain of the hERG1a splice variant, and thus putatively specific for the latter isoform. The effects of these siRNAs on both RNA and protein levels were analysed 24 and 48 hours after transfection, on HEK 293 cells stably transfected with the hERG1a and the hERG1b isoforms, respectively. One siRNA turned out to specifically inhibit hERG1A protein expression by 60%, without affecting hERG1B protein level. The same effect was observed in neuroblastoma cells, that endogenously express both the hERG1A and the hERG1B isoforms. It is possible to conclude that RNAi can be used to dissect the function of different splice variants of ion channels encoding genes in different physiological and pathological situations. 1 Meister G and Tuschl T, Nature, 431, 343-349, 2004 Hannon GJ and Rossi JJ, Nature, 431, 371-378, 2004 3 Arcangeli A In: The hERG cardiac potassium channel: structure, function, and long QT syndrome. Wiley, Chichester (Novartis Foundation Symposium 266), pp.225-234, 2005. 4 Crociani O et al., J.Biol.Chem.,278,2947-55,2003. 2