classification of Alteromonas spp. using multilocus sequence
advertisement

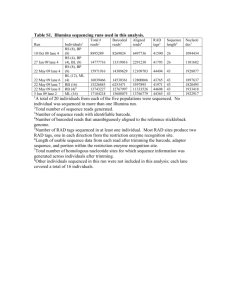
Supplementary material Updating the taxonomic toolbox: classification of Alteromonas spp. using multilocus sequence analysis and MALDI-TOF mass spectrometry Hooi Jun Nga, Hayden K. Webba, François Malherbea, Russell J. Crawforda, Henry Buttb, Rachel Knightb, Valery V. Mikhailovc and Elena P. Ivanovaa, * a Faculty of Life and Social Sciences, Swinburne University of Technology, PO Box 218, Hawthorn, Vic 3122, Australia b c Bioscreen, Bio21 Institute, The University of Melbourne, Vic 3010, Australia Pacific Institute of Bioorganic Chemistry, Far-Eastern Branch of the Russian Academy of Sciences, Vladivostok 690022, Russian Federation *Corresponding author: Tel: +61-3-9214-5137. Fax: +61-3-9214-5050. E-mail: eivanova@swin.edu.au Fig. S1 Agarose gel electrophoresis of PCR products from five house-keeping genes (dnak, sucC, rpoB, gyrB, and rpoD). Lane M, TrackIt™ 1 Kb Plus DNA Ladder (Invitrogen, USA); Lane 1, A. addita; Lane 2, A. genovensis; Lane 3, A. hispanica; Lane 4, A. litorea; Lane 5, A. macleodii; Lane 6, A. marina; Lane 7; A. simiduii; Lane 8, A. stellipolaris; Lane 9, A. tagae; Lane 10, Pseudoalteromonas translucida; Lane 11, Shewanella colwelliana; Lane 12, Marinomonas communis; Lane 13, Salinimonas chungwhensis; Lane 14, Aestuariibacter aggregatus; Lane 15, Glaciecola mesophila. Fig. S2 Phylogenetic analyses of the nucleotide sequences of each of the five markers (gyrB, dnaK, rpoB, sucC, rpoD) genes used for MLSA of Alteromonas spp. The trees were calculated using Neighbour Joining method. Numbers at branching points are percentage bootstrap values based on 1000 replications, with only values > 50% shown. Scale bar represents 0.01/ 0.02 substitutions per nucleotide position. Fig. S3 Phylogenetic analysis of the concatenated amino acid sequences (DnaK, SucC, RpoB, GyrB, and RpoD). Numbers at branching points are percentage bootstrap values based on 1000 replications, with only values > 50% shown. Scale bar represents 0.005 substitutions per position. Fig. S4 Main Spectra Library (MSP) dendrogram of MALDI-TOF mass spectral profiles from nine Alteromonas species and type strains of closely related species generated by the MALDI Biotyper 3.0 software. Distance is displayed in relative units.