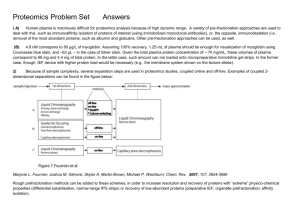
Protein Structure
advertisement

Chapter 5: Outline Amino Acids Amino acid classes Bioactive AA Modified AA Peptides Proteins (We are here) Protein structure Fibrous proteins Globular proteins Stereoisomers Titration of AA AA reactions 5P2-1 Protein Function 1. Catalysis 2. Structure 3. Movement 4. Defense 5. Regulation 6. Transport 7. Storage 8. Stress Response 5P2-2 Proteins by Shape-1 Fibrous proteins exist as long stranded molecules: Eg. Silk, collagen, wool. A collagen segment in space-filling mode illustrates this point. Red spheres represent oxygen, grey carbon, and blue nitrogen 5P2-3 Proteins by Shape-2 Globular proteins have somewhat spherical shapes. Most enzymes are globular. Eg. myoglobin, hemoglobin. Myoglobin in space-filling mode is the chosen example. 5P2-4 Proteins by Composition Simple Contain only amino acids Conjugated simple protein (apoprotein) prostetic group (nonprotein) glycoproteins lipoproteins metaloproteins etc. Holoprotein 5P2-5 Four Levels of Protein Structure Primary, 1o the amino acid sequence Secondary, 2o 3-D arrangement of backbone atoms in space Tertiary, 3o 3-D arrangement of all the atoms in space Quaternary, 4o 3-D arrangement of subunit chains 5P2-6 Determining Primary Structure 1. Hydrolyze protein with hot 6M HCl. Identify AA and % of each. Usually done by chromatography 2. Identify the N-term and C-term AAs C-term via carboxypeptidase N-term via Sanger’s Reagent, DNFB 2,4-dinitrofluorobenzene Often step 2 can be skipped today. 5P2-7 Det. Primary Structure: 2 3. Selectively fragment large proteins into smaller ones. Eg. Tripsin: cleave to leave Arg or Lys as C-term AA Eg. Chymotrypsin: cleave to leave Tyr or Trp or Phe as C-term AA Eg. Cyanogen bromide cleaves at internal Met leaving Met as C-term homoserine lactone 5P2-8 Det. Primary Structure: 3 4. Determine AA sequence of peptides with AA sequencer using Edman’s reagent: phenyl isothiocyanate which reacts with the N-term AA See the next slide 5P2-9 Det. Primary Structure: 3b + H3N 1 2 - COO 3 protein Edman’s reagent N C S S NH C NH 1 2 3 Thiazoline derivative N CH R1 NH C S C O C C S NH + H3N + 2 3 - COO O aqueous acid RAR CH R1 N - COO Phenylthiohydantoin (PTH) derivative of N-term AA 5P2-10 Det. Primary Structure: 4 5. Reassemble peptide fragments from step 3 to give protein. An example follows on the next slide. 5P2-11 Det. Primary Structure: 4b A twelve AA peptide was hydrolyzed. Trypsin hydrolysis: Leu-Ser-Tyr-Gly-Ile-Arg One is Thr-Ala-Met-Phe-Val-Lys C-term Chymotrypsin hydrolysis Val-Lys-Leu-Ser-Tyr Lys is internal! Gly-Ile-Arg Thr-Ala-Met-Phe Deduce the AA sequence 5P2-12 Det. Primary Structure: 4c Keeping in mind the N-term AA and overlaping the sequences properly gives: Tr Leu-Ser-Tyr-Gly-Ile-Arg Ct Gly-Ile-Arg Ct Val-Lys-Leu-Ser-Tyr Tr Thr-Ala-Met-Phe-Val-Lys Ct Thr-Ala-Met-Phe The complete sequence is: Thr-Ala-Met-Phe-Val-Lys-Leu-Ser-Tyr-Gly-Ile-Arg 5P2-13 Secondary Structure The two very important secondary structures of proteins are: a-helix b-pleated sheet Both depend on hydrogen bonding between the amide H and the carbonyl O further down the chain or on a parallel chain. 5P2-14 a Helix: Peptide w Hbonds First six C=O to N hydrogen bonds shown 5P2-15 b Sheet: stick form Protein G H bonds in dotted red-blue H bonds shown in dotted red-blue Chain segment 1 Seg 2 Seg 3 Chain 1 Seg 4 5P2-16 B Sheet: Lewis Structure N-term C-term CH C O H N HC O C N H CH C O H N HC H N C O CH N H O C HC H N C O C-term N-term Antiparallel sheet N-term CH C O H N HC O C N H CH C O H N C-term N-term CH C O H N HC O C N H CH C O H N C-term Parallel sheet 5P2-17 Supersecondary Structure Reverse turns in a protein chain allow helices and sheets to align side-by-side Common AA found at turns are: glycine: small size allows a turn proline: geometry favors a turn 5P2-18 Supersecondary Structure: 2 Combinations of a helix and b sheet. bab aa b meander 5P2-19 Tertiary Structure The configuration of all the atoms in the protein chain: side chains prosthetic groups helical and pleated sheet regions 5P2-20 Tertiary Structure: 2 Protein folding attractions: 1. Noncovalent forces a. Inter and intrachain H bonding b. Hydrophobic interactions c. Electrostatic attractions + to - ionic attraction d. Complexation with metal ions e. Ion-dipole 2. Covalent disulfide bridges 5P2-21 Tertiary interactions: diag. disulfide + NH3 O O C Polypeptide Chain CH2 S S CH2 CH3 CH3 HO CH2 CH2 OH metal coord’n Mg2+ H3C CH CH3 O O H3C CH CH3 O H C ionic Ion-dipole hydrophobic + NH3 O O C H bonds or dipole 5P2-22 Domains Domains are common structural units within the protein that bind an ion or small molecule. 5P2-23 Quaternary Structure-1 Quaternary structure is the result of noncovalent interactions between two or more protein chains. Oligomers are multisubunit proteins with all or some identical subunits. The subunits are called protomers. two subunits are called dimers four subunits are called tetramers 5P2-24 Quaternary Structure-2 If a change in structure on one chain causes changes in structure at another site, the protein is said to be allosteric. Many enzymes exhibit allosteric control features. Hemoglobin is a classic example of an allosteric protein. 5P2-25 Denaturation -loss of protein structure, not 1o. 1. Strong acid or base 2. Organic solvents 3. Detergents 4. Reducing agents 5. Salt concentrations 6. Heavy metal ions 7. Temperature changes 8. Mechanical stress o 2 o 4 , but 5P2-26 Denaturation-2 Denaturing destroys the physiological function of the protein. Function may be restored if the correct conditions for the protein function are restored. But! Cooling a hardboiled egg does not restore protein function!! 5P2-27 Fibrous Proteins Fibrous proteins have a high concentration of a-helix or b-sheet. Most are structural proteins. Examples include: a-keratin collagen silk fibroin 5P2-28 Globular Proteins Usually bind substrates within a hydrophobic cleft in the structure. Myoglobin and hemoglobin are typical examples of globular proteins. Both are hemoproteins and each is involved in oxygen metabolism. 5P2-29 Myoglobin: o 2 and o 3 aspects Globular myoglobin has 153 AA arranged in eight a-helical regions labeled A-H. The prosthetic heme group is necessary for its function, oxygen storage in mammalian muscle tissue. His E7 and F8 are important for locating the heme group within the protein and for binding oxygen. A representation of myoglobin follows with the helical regions shown as ribbons. 5P2-30 Myoglobin: 2o and 3o aspects Some helical regions Heme group with iron (orange) at the center 5P2-31 The Heme Group - - CH2CH2COO OOC CH2CH2 N of His F8 binds CH3 H3C to N N fifth site on the iron. Fe(II) Pyrrole ring H2C N N CH CH3 His E7 acts as a ”gate” CH3 CH CH2 for oxygen. 5P2-32 Binding Site for Heme Lower His bonds covalently to iron(II) Oxygen coordinates to sixth site on iron and the upper His acts as a “gate” for the oxygen. N N H O N O N Fe N N N N 5P2-33 Hemoglobin A tetrameric protein two a-chains (141 AA) two b-chains (146 AA) four heme units, one in each chain Oxygen binds to heme in hemoglobin cooperatively: as one O2 is bound, it becomes easier for the next to bind. Lengthy segments of the a and b chains homologous to myoglobin. 5P2-34 Hemoglobin: ribbons + hemes Each chain is in ribbon form and color coded. The heme groups are in space filling form 5P2-35 Oxygen Binding Curves Oxygen bonds differently to hemoglobin and myoglobin. Myoglobin shows normal behavior while hemoglobinn shows cooperative behavior. Each oxygen added to a heme makes additon of the next one easier. The myoglobin curve is hyperbolic. The hemoglobin curve is sigmoidal. 5P2-36 Oxygen Binding Curves-2 5P2-37 The Bohr Effect (H+ and Hb) Lungs: pH higher than in actively metabolizing tissue. (Low H+). Hb binds oxygen and + releases H . Muscle at Work: + pH lower (H product of metabolism). Hb releases oxygen and binds H+. HbO2 + H+ + CO2 metabolism in lungs O2 + H+-Hb-CO2 5P2-38