SPECTROPHOTOMETRY
advertisement

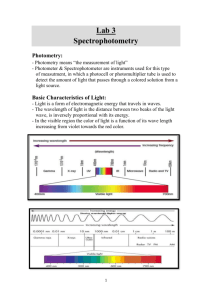
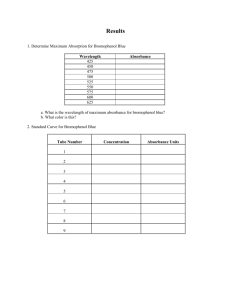
SPECTROPHOTOMETRY Spectrophotometry • Determines concentration of a substance in solution – Measures light absorbed by solution at a specific wavelength Spectrophotometry • One of the simplest and most widely used methods to determine the amount of protein or nucleic acid present in a given solution Spectrophotometry • Proteins do not absorb in visible wavelength region unless they have a prosthetic group (e.g., Fe2+), or an unnatural amino acid Spectrophotometry • The amino acids tryptophan, tyrosine & cytosine absorb light in the UV wavelength • Aromatic rings in the bases of nucleic acids also absorb light in the UV range Spectrophotometry • Visible region: low energy electronic transition due to: a. Compounds containing transition metals b. Large aromatic structures & conjugated double bond systems (vitamin A, retinal, heme) • UV region (200-400 nm): a. Small conjugated ring systems (Phe, Tyr, Trp) Spectrophotometry Io I A = 0.012 l Lamp Monochromator Detector Cuvette Spectrophotometers • • • • Light source (Lamp) Optical filters or prism Tube or cuvette Photocell or photomultiplier tube Light source (Lamp) • Visible region = tungsten or tungstenhalogen • UV light = deuterium or hydrogen lamp Optical filters/prisms • To limit light to a certain wavelength • Monochromator can isolate a specific wavelength of white light and allow it to pass through the solution being analyzed Tubes or cuvettes • Visible range = glass cuvette • UV range = quartz cuvette Photocell • To detect transmitted light Spectrophotometry • Beer-Lambert’s Law lo Where: g Io = cl I Io = intensity of incident light I = intensity of transmitted light = molar extinction coefficient c = concentration of the absorbing species (mol/L) l = path length of the light-absorbing sample (cm) Beer-Lambert’s Law • The fraction of the incident light absorbed by a solution at a given wavelength is related to a. thickness of the absorbing layer (path length) and b. concentration of the absorbing species Visible region wavelength Color Ultraviolet Violet Blue Green Yellow Orange Red Infrared Wavelength (nm) 400 and under 400 - 450 450 - 500 500 - 570 570 - 590 590 - 620 620 - 650 750 & over Beer-Lambert’s Law • Concentration amount of light absorbed A = abc = log(100/%T) Where A = absorbance a = absorptivity of the compound under standard conditions b = light path of the solution c = concentration of the compound %T = percent transmittance Beer-Lambert’s Law • Absorbance A = K x C = Log10Io I Where: Io = amount of light absorbed by the solution expressed as absorbance or optical density K = constant C = concentration of the substance Transmittance • Defined as the ratio of the intensity of light emerging from the solution (I) to that of incident light entering (Io) T=I Io Io I Transmittance • Inversely related to the concentration of the solution and is expressed in % % T = 1 x 100 Io Transmittance • 100% transmittance means no light is absorbed by the solution so that incident light is 100% transmitted Absorbance & Transmittance • Absorbance concentration • Transmittance 1/ to concentration and absorbance Sample Problem • Cytosine has a molar extinction coefficient of 6 x 103 mol-1 cm-1 at 270 nm at pH 7. Calculate absorbance of 1 x 10-3 M cytosine solution in 1mm cell at 270 nm A = Log I0 = lc I Sample Problem • Solution: 1. A = lc = (6 x 103)x (0.1) x (1 x 10-3) = 6 x 10-1 = 0.6 (O.D.) O.D. between 0.1 and 2 are most reliable Spectrophotometry • Clinical applications: 1. Aromatic amino acids have characteristic strong absorbance of light at a wavelength of 280 nm ex. Tryptophan & tyrosine Calculation Cu = Cs x A(u) x D A(s) Where: Cs = concentration of standard Cu = concentration of unknown A(s) = absorbance of standard A(u) = absorbance of unknown D = dilution factor Calibration Curve Glucose Std. Concn. Absorba nce 60 mg% 0.2 120 mg% U 0.4 0.5 Absorbance Glucose Standard Calibration Curve 1.2 1 0.8 0.6 0.4 0.2 Linear ( ) 0 180 mg% 0.6 60 120 180 Mg% glucose 200 Colorimetric determination of reducing sugars • Dinitrosalicylate • Potassium ferric hexacyanid (Prussian blue) • Nelson-Somogyi (molybdenum blue) DNS method • Developed by Sumner & Sisler (1944) and modified by Miller (1959) • Based on reduction of sugars by DNS under alkaline conditions to yield 3amino-5-nitrosalicylate (brown color) DNS method • Measured at 540 nm • Quantity of reducing sugar is extrapolated from a calibration curve prepared with D-glucose • Amylase-catalyzed reactions are typically buffered at pH5 using acetate or citrate DNS method • Amylase-catalyzed reactions are typically buffered at pH 5 using acetate or citrate • Citrate may interfere with DNS color development Principle • Carbohydrates are essentially aldehydes or ketones that contain multiple hydroxyl (-OH) groups • Monosaccharides can be aldoses (glucose) or ketoses (fructose Principle • Both aldoses & ketoses occur in equilibrium between the open-chain forms and cyclic forms (chain lengths of C4) • These are generated by bond formation between one of the (-OH) groups of the sugar chain with the C of the aldehyde or keto group to form a hemiacetal bond. Principle Principle Principle Principle • When salivary amylase is added to starch, a hydrolysis reaction is initiated in which water breaks bonds, releasing maltose Principle • DNS tests for the presence of free carbonyl groups (C=O), the so-called reducing sugars • Involves oxidation of the aldehyde functional groups in glucose and the ketone functional groups in fructose Principle • Simultaneously, 3,5 DNS is reduced to 3-amino, 5 nitrosalicylic acid under alkaline conditions • As hydrolysis proceeds, more reducing sugar will be available to react with the 3,5 DNS Principle oxidation Aldehyde group carboxyl group reduction 3,5 Dinitrosalicylic 3-amino, 5 nitrosalicylic Standard Absorbance Curve • Done by reacting know concentration of glucose with DNS then determining absorbance at 540 nm • Plot absorbance vs. glucose concentration Absorbance • Absorbance corresponds to 0.1 ml of test = x mg of glucose 10 ml contains = x (10 mg of glucose) 0.1 = % of reducing sugars