(A) HLA-B*4402 and
advertisement
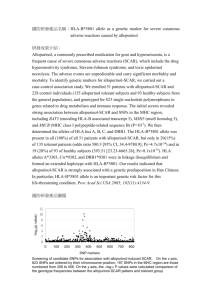
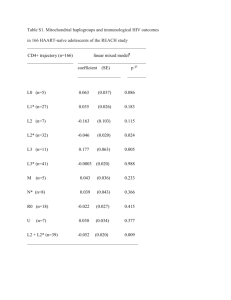
Polymorphism in position 156 influences the peptide repertoire of HLA-B*4402 and B-4403 molecules Hernando Escobar, Eduardo Reyes-Vargas, David Crockett, Peter Jensen, and Julio C. Delgado ARUP Institute for Clinical and Experimental Pathology, Department of Pathology, University of Utah School of Medicine, Salt Lake City, Utah Strong CTL-mediated allogeneic reactions have been reported between HLA-B*4402 and –B*4403 alleles. These alleles differ only by a single residue located in position 156 of the α chain, with HLA- B*4402 expressing Asp and HLA-B*4403 expressing Leu. Position 156 influences the conformation of pocket D and E of the peptide binding groove of HLA class I molecules but has little influence on the specificity of peptide anchor residues P2 and P9. This study was performed to investigate the influence of position 156 in the repertoire peptides presented by these two HLA-B44 subtypes. HLA-B44-peptide complexes were separated by immuno-affinity columns from whole cell lysates of K562 cells expressing HLA-B*4402 or HLA-B*4403. Peptide pools were obtained by acid elution followed by HPLC fractionation and sequencing by CID and TOF MS/MS. We found that over 2/3 of peptides eluted from HLA- B*4402 differ from those eluted from HLA-B*4403. Close analysis of the peptide binding motif of non-over-lapping peptide sequences of these 2 molecules revealed that in the non-anchor position P7, HLA-B*4402 prefers polar, uncharged and basic residues such as Gly, Gln or His, whereas HLA*B4403 shows preference for non-polar residues Leu, Ile or Val. No differences were observed in the specificity of residues located in anchor positions P2 and P9. Position 156 has direct effect on the specificity of the residue located in position P7 of the peptide binding motif of HLAB*4402 and HLA-B*4403 alleles. Given that P7 position is important for T-cell receptor interaction and subsequent T-cell activation, we suggest that the polymorphism in position156 between HLA-B*4402 and HLA-B*4403 alleles might play an important role in the alloreactivity between these 2 alleles. Eluted peptides from K562 cells expressing HLA-B*4402 or B*4403 were processed as described in Escobar, H. a. et al., J. Immunol. 2008; 181,4874-4882. In brief, MHC molecules were immunopurified from whole cell lysates and peptides were acid-eluted and fractionated by HPLC. Peptides fractions were analyzed by LC/MS/MS using an Agilent 6510 Q-TOF (Agilent Technologies, Santa Clara, CA) equipped with nanospray ChipCube source and Agilent 1200 HPLC system. Peptide separation was performed on a C-18SB-ZX Chip (40nL trap, 150mm x 43mm, 5mm particles). Data acquisition was performed with MassHunter QTOF Acquisition software B.02 with an acquisition rate of 3 scans/s followed by MS/MS scans of 3 most intense ions. Exclusion was set for 12s after two consecutive MS/MS scans of the precursor ion. TOF mass analyzer was calibrated and tuned prior to each set of samples with mass accuracy < 1 ppm. The acquired MS/MS spectra were searched using Spectrum Mill MS Proteomics Workbench Rev A.03.03.075 (Agilent) against SwissProt human or mouse database. Spectra were validated automatically and filtered with the following settings: minimum score 7.0, minimum 60 percent scored peak intensity SPI, minimum unmatched ions 15 of 30 total ions, maximum precursor error 20ppm. Sequences were then manually validated after the initial database search as reported in Delgado, J.C. et al Immunogenetics, 61: 241–246. b. Using large-scale sequencing of eluted endogenous peptides we identified 52 peptides from HLA- B*4402 and MHC class I molecules are highly polymorphic structures that bind peptide antigens for 62 HLA-B*4403. As expected, both peptide repertoires originate from intracellular compartments and have presentation to T cells and generate an immune response against pathogens. Several similar function (Fig 2). Approximately 50% of B*4402-derived peptides are shared with B*4403, whereas only MHC class I molecules can be classified in 10-12 major HLA class I supertypes which are 37% of B*4403-derived peptides are shared with B*4402. Analysis of peptide sequences confirmed previous characterized by specific anchor binding motifs. The HLA-B44 supertype is present in reports showing a dominant anchor Glu residue in P2 position and the presence of aromatic or hydrophobic most human populations where HLA-B*4402 is common in Caucasian and HLA-B*4403 is residues as Phe, Tyr and Leu in the last position (Fig 3). Analysis of non-common peptide sequences revealed common in African and Asian populations. Between this two B44 alleles there is only a that in position P7, HLA-B*4402 prefers polar, uncharged and basic residues such as Gly, Gln or His, whereas single residue difference located in position 156 (Asp in HLA- B*4402 and Leu in HLA- HLA*B4403 shows preference for non-polar residues Leu, Ile or Val. B*4403). Although position 156 is buried in the antigen-binding cleft of MHC class I molecule, its effect on T cell recognition is crucial for allograft rejection in clinical transplantation (Macdonald WA, et al. J. Exp. Med, 2003; Ely LK, et al. J. Immunol, 2005). As seen in Fig 1, position 156 is located in the floor of the binding cleft and affects pockets Figure 3. Logos displaying peptide-binding motifs of non-common peptides found in: (a) HLA-B*4402 and (b) HLA-B*4403. Residues acidic (red), basic (blue), hydrophobic (black), neutral (green) and aromatic (purple) are illustrated. Dominant P2 position occupied by Glu has been removed to show in better detail the frequencies of the remaining positions. D and E of MHC class I molecules and has a strong effect on T cell recognition (Herman JV et al. Tissue Antigens, 1999) a. 1. Our results demonstrate that position 156 in has direct effect on the specificity of the residue located in position P7 of the peptide binding HLA-B*4402 and HLAB*4403 molecules. b. 2. Previous studies have shown that P7 position has a potential T-cell receptor interaction and subsequent T-cell activation (Archbold JK, et al. JEM, 2009 ). Thus, the polymorphism in position 156 might play an important role in the observed alloreactivity between HLA-B*4402 and HLA-B*4403 alleles. Figure 1. Top side view of peptide binding cleft of (a) HLA-B*4402 and (b) HLA–B*4403 showing Asp and Leu residues in position 156, respectively, and its influence in pockets D and E. Figure 2. Gene Ontology classification of identified peptides from (A) HLA-B*4402 and (B) HLA-B*4403. 3. In-vitro binding assays are currently being performed using sets of peptides with variation of residues in position P7 to demonstrate the effect of this position in the binding to HLA-B*4402 and HLA-B*4403 molecules