feb2s0014579315001878-sup-m0005
advertisement

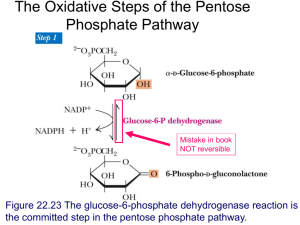
1. Primer sequences used in this study for cloning and mutagenesis Target GBE VvGBE Wild type EGBE F/R 5’-Sequence of Oligonucleotides-3’ Restriction enzyme F TACGCGGTCGCATATGAGGATTGTGTTCAC NdeI R F CCTAATCGCCTGCAGTTAGACCAGCTTGTAG CAGGAAGACACATATGTCCGATCGTATC PstI NdeI R CAATGGCGAAAGCTTTCATTCTGCCTCC HindⅢ F GCCCCGTCCATATGCTTGACCCGTTG NdeI R GCGGACGAACTGCAGTCACCGGTCCTCG PstI F R F R F R F R F R F R AGTATGACGATCATATGACGCCGAAAACCATG CATGCCTGCTCGAGGACCAGCTTGTAGAAC GTCACGTACCATATGGCTCGTTACCAGTGGC CATGCCTGCTCGAGGACCAGCTTGTAGAAC CACCATCACCATATGAGGATTGTGTTCAC CATGGTTTTCTGCAGGTGGAGATCGTC GCTATTATCTCTGCAGACTCACCTGCGCC CAATGGCGAAAGCTTTCATTCTGCCTCC CACCATCACCATATGAGGATTGTGTTCAC GCCACTGGTACTGCAGGTGATCGTACG CCTTATGCCTTTCTGCAGCAAATGCGCC GATTACGCCAAGCTCGAGTTCTGCCTCCC NdeI XhoI NdeI XhoI NdeI PstI PstI HindⅢ NdeI PstI PstI XhoI DgGBE Truncation mutants VvGBE-N1 VvGBE-N VvGBE N1 EGBE Swapping mutants N VvGBE EGBE 2. The characteristics of vectors and recombinant plasmids used in this study Strains and Vectors Strains Characteristics Vibrio vulnificus MO6-24/O Escherichia coli MC1061 Deinococcus geothermalis DSM 11300 p6xHis119 Vectors p6xHTKNd pTKNd6xH [1] [2] [3] ∙ Pblma, constitutive, Amr ∙ His-tag (N-terminal) ∙ Pblma, constitutive, Kmr ∙ His-tag (N, C-terminal) ∙ Pblma, constitutive, Kmr ∙ His-tag (C-terminal) Recombinant plasmids Recombinant enzyme Wild type Cloning vector VvGBE EGBE DgGBE ∙ p6xHis119 vector, Amr ∙ p6xHis119 vector, Amr ∙ p6xHis119 vector, Amr VvGBE-N1 (N1_t) ∙ p6xHTKNd vector, Kmr ∙ 129-residue deletion at the N-terminal of VvGBE VvGBE-N (N_t) ∙ p6xHTKNd vector, Kmr ∙ 240-residue deletion at the N-terminal of VvGBE N1 swapping (N1_s) ∙ p6xHis119 vector, Amr ∙ N1 of VvGBE replaced counterpart of E.coli GBE N swapping (N_s) ∙ pTKNd6xH vector, Kmr ∙ N domain of VvGBE replaced the counterpart of E.coli GBE Truncation mutants Swapping mutants 3. Sequences used in the phylogenetic analysis All amino acid sequences and conserved domain searches of known GBEs were obtained from the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov): Aquifex aeolicus VF5 (gi.15606119), Bacillus cereus (674471943), Bacillus subtilis 168 (16080150), Deinococcus geothermalis DSM 11300 (94555367), Deinococcus radiodurans R1 (15806848), Escherichia coli K-12 MG1655 (16131306), Mycobacterium tuberculosis (631464201), Salmonella enterica serovar Typhi Ty21a (485084115), Thermococcus kodakarensis KOD1 (74502442), Thermus thermophiles (504323830), Vibrio cholerae (669407972), Vibrio parahaemolyticus (505126546), and Vibrio vulnificus MO6-24/O (319933217). HPAEC analysis of glycogen resulting from adding G2 by V.vulnificus MO6-24/O 4. Glycogen in Vibrio vulnificus MO6-24/O 20 G1-G7 std after 2 h (control) after 2 h + Isoamylase Response (nC) 15 10 5 0 G4 G5 G7 G6 G3 G4 G5 G6 HPAEC analysis of glycogen resulting from adding G2 by V.vulnificus MO6-24/O -5 20 10 30 40 50 G1-G7 std Retention time (min) after 4 h (control) after 4 h + Isoamylase 15 Response (nC) 20 10 5 0 HPAEC analysis of glycogen resulting from adding G2 by V.vulnificus MO6-24/O -5 Response (nC) 30 10 20 30 40 50 G1-G7 std Retention time after (min)6 h (control) after 6 h + Isoamylase 20 10 0 10 20 30 40 50 Retention time (min) Culture condition: Cells were cultured with Luria-Bertani medium at 37°C with shaking. Glycogen extraction and side-chain distribution analysis: After cell growth reach late exponential phase, 1.0% maltose (w/v in final) was added. Cells were harvested by centrifugation by 2 h time intervals after c-source addition. Glycogen was extracted and analyzed as described by Park et al. previously (J. Bacteriol. 193(10):2517-2526, 2011). Control means that glycogen was injected directly without debranching by isoamylase. References for supplementary materials 1. Park, J. H., Cho, Y.-J., Chun, J., Seok, Y.-J., Lee, J. K., Kim, K.-S., Lee, K.-H., Park, S.-J. & Choi, S. H. (2011) Complete Genome Sequence of Vibrio vulnificus M06-24/O, Journal of Bacteriology. 2. Park, J.-T., Shim, J.-H., Tran, P. L., Hong, I.-H., Yong, H.-U., Oktavina, E. F., Nguyen, H. D., Kim, J.-W., Lee, T. S. & Park, S.-H. (2011) Role of maltose enzymes in glycogen synthesis by Escherichia coli, Journal of bacteriology. 193, 2517-2526. 3. Makarova, K. S., Omelchenko, M. V., Gaidamakova, E. K., Matrosova, V. Y., Vasilenko, A., Zhai, M., Lapidus, A., Copeland, A., Kim, E. & Land, M. (2007) Deinococcus geothermalis: the pool of extreme radiation resistance genes shrinks, PLoS One. 2, e955.