feb212044-sup-0001-Supinfo
advertisement

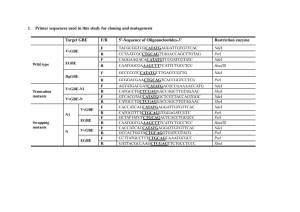
Supplementary Methods Construction of plasmids All of the primers used in this study are listed in supplementary Table 1. To construct the expression plasmid for GST-TbPex14p or TbPex5p-His, the DNA fragment encoding TbPex14p or TbPex5p were inserted into pGEX-6P-1 (GE Healthcare) or pET24b (Novagen, Madison, WI), respectively. A fragment with the 1.1-kb TbPEX14 coding region was amplified with the primer set TbPEX14-Bam-Fw/TbPEX14-Not-Rv using genomic DNA as the template. Similarly, the 2.0-kb TbPEX5 coding region was amplified with the primers TbPEX5-Nde-Fw/TbPEX5-Xho-Rv. The PCR products were digested with BamHI/NotI or NdeI/XhoI, then ligated into the BamHI–NotI site of pGEX-6P-1 or NdeI–XhoI site of pET24b, respectively, to yield pGEX-6P-1/TbPEX14 and pET24b/TbPEX5. The expression plasmids for TbPex5p(a.a.47–60), TbPex5p(a.a.196–208) and TbPex5p(a.a.310–333), which contained the deduced Pex14p binding motifs fused with a His-tagged lipoyl domain at the N-terminus, were constructed as follow. First, the 0.9-kb TbPex5p(a.a.47–333) coding region was amplified with the primer set Tb5motif1NdeI-Fw/Tb5motif3BamHI-Rv using pET24b/TbPEX5 as the template. The PCR product was digested with NdeI and BamHI, then ligated into the NdeI–BamHI site of pOPHLT to yield pOPHLT/TbPEX5(a.a.47–333). Then the 3.3-kb fragments containing the TbPEX5(a.a.47–60) or TbPEX5(a.a.310–333) region was amplified by inverse PCR using the primer sets Tb5M1inv-Fw/Tb5M1inv-Rv or Tb5M3inv-Fw/Tb5M3inv-Rv using pOPHLT/TbPEX5(a.a.47–333) as the template. This fragment was self-ligated using T4 Polynucleotide Kinase (Toyobo) and Ligation high (Toyobo) to form pOPHLT/TbPEX5(a.a.47–60) 1 and pOPHLT/TbPEX5(a.a.310–333). pOPHLT/TbPEX5(a.a.196–208) and pOPHLT/TbPEX5(a.a.310–333) were constructed from pOPHLT/TbPEX5(a.a.47–333) according to the same procedure described above. The DNA fragment encoding TbPex14p (a.a.23–71) was amplified Tb14domain-Nde-Fw/Tb14domain-Bam-Rv using with the primer pGEX-6P-1/TbPEX14 set as the template. The PCR product was digested with NdeI and BamHI, then ligated with the NdeI/BamHI fragment of pOPHBL to yield pOPHBL/TbPEX14(a.a.23–71). All of the cloned inserts were confirmed using the Big Dye V.3.1 Terminator Kit (Applied Biosystems) in an ABI PRISM 3130 sequencer (Applied Biosystems). Purification of TbPex5p-His and GST-TbPex14p TbPex5p-His6 was overexpressed using E. coli BL21(DE3)pLysS cells (Novagen, Darmstadt, Germany) as the host. The E. coli cells harboring pET-24b/TbPEX5 were grown at 37°C in LB media containing 50 µg/ml kanamycin. At a cell density of 0.7 (OD600), protein expression was induced with 0.5 mM IPTG for 4 h at 37°C. The cells were harvested by centrifugation at 4,000 x g for 20 min, resuspended in 3.5 ml of buffer A (50 mM Hepes, pH7.5, 150 mM NaCl, 4 mM β-mercaptoethanol, 1% Triton X-100, 5% glycerol and 5 mM imidazole) and disrupted 20 times for 20 s in an ice bath using an Astrason XL-2020 ultrasonic processor (Misonix Inc., Farmingdale, NY). The lysate was centrifuged at 20,000 x g for 20 min and TbPex5p-His in the supernatant was immediately applied to 250 µl of cOmplete His-Tag Purification resin (Roche Diagnostics GmbH, Mannheim, Germany) equilibrated with the lysis buffer A. After extensive washing, the TbPex5p-His was eluted with buffer A containing 250 mM imidazole. The TbPex5p-His mutants were purified with the same procedure. 2 GST-TbPex14p was also overexpressed using E. coli BL21(DE3)pLysS cells as the host. The E. coli cells harboring pGEX-6P-1/TbPEX14 were grown at 37°C in LB media containing 100 µg/ml ampicillin. At a cell density of 0.6 (OD600) protein expression was induced with 0.5 mM IPTG for 10 h at 20°C. The cells were harvested by centrifugation at 4,000 x g for 20 min, resuspended in 35 mL of buffer B (50 mM Hepes, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 1% Triton X-100 and 10% glycerol) and disrupted 20 times for 20 s in an ice bath with an Astrason XL-2020 ultrasonic processor. The lysate was centrifuged at 20,000 x g for 20 min and the GST-TbPex14p in the supernatant was immediately applied to 10 ml of glutathione-Sepharose 4B resin (GE Healthcare, Little Chalfont, UK) equilibrated with buffer B. After extensive washing, the GST-TbPex14p was eluted with buffer B containing 20 mM glutathione. Purification of His-Lipoyl-TbPex5p(a.a.47–60), -TbPex5p(a.a.196–208), -TbPex5p(a.a.310–333), -TbPex14p (a.a.23–71), and TbPex14p (a.a.23–71) E. coli C41(DE3)RIPL cells were pOPHLT/TbPEX5(a.a.47–60), pOPHLT/TbPEX5(a.a.310–333) transformed with the plasmids, pOPHLT/TbPEX5(a.a.196–208), or pOPHLT/TbPEX14(a.a.23–71) according to standard procedures. The E. coli cells harboring these plasmids were grown at 37°C in LB media containing 0.1 mg/mL ampicillin. At a cell density of 0.5 (OD600) protein expression was induced with 0.3 mM IPTG for 3 h at 37°C. The cells were harvested by centrifugation at 3,765 x g for 10 min, resuspended in buffer C (20 mM Tris-HCl, pH 7.5, 100 mM NaCl and 1 mM dithiothreitol) and lysed by sonication on ice. The lysate was centrifuged at 100,000 x g for 20 min, then the supernatant was passed through the 3 0.45 µm filter. The supernatant was applied to HisTrap resin (GE Healthcare) equilibrated with lysis buffer C. After extensive washing, the recombinant protein was eluted using a linear gradient with an increasing imidazole concentration (0 to 500 mM) in elution buffer (20 mM Tris-HCl, pH 7.5, 100 mM NaCl, and 500 mM imidazole). The recombinant proteins were further purified by anion exchange chromatography (by) with an HiTrap Q HP column (GE Healthcare) followed by gel filtration chromatography with Superdex 75 16/60 (GE Healthcare). TEV protease was used for His-Lipoyl-TbPex14p (a.a.23–71). TbPex14p (a.a.23–71) was purified by gel filtration chromatography. 4 Supplementary Table 1 List of oligonucleotide primer sequences. TbPEX14-Bam-Fw 5’-AAATTTATAGGATCCATGTCTTTGCTGCTGTCGGG-3’ TbPEX14-Not-Rv 5’-AAATTTATAGCGGCCGCTCAAGCTGCCTCGCCGCCAA-3’ underbar shows BamHI and NotI site, respectively TbPEX5-Nde-Fw 5’-AAATTTATACATATGATGGACTGCGGCGCCGG-3’ TbPEX5-Xho-Rv 5’-AAATTTATACTCGAGCCGGCGCCGCAGTCCAT-3’ underbar shows NdeI and XhoI site, respectively Tb5W52A/F56A-Fw 5'-AGCACGCTGCTGCCCATCAACATCACCAC-3' Tb5W52A/F56A-Rv 5'-GAGCCGCGTCTTCCATGGGCCCGGTGGG-3' Tb5W200A/Y204A-Fw 5'-AGGATGCCAAGGATGTTGAGGTGCATACG-3' Tb5W200A/Y204A-Rv 5'-GTCCCGCCTCCGCCTGGTGCAATTTCTC-3' Tb5W318A/Y322A-Fw 5'-AGGAAGCCGCACAAATGCAGGCCATGCAG-3' Tb5W318A/Y322A-Rv 5'-GTGCCGCCTGCTCAACGTCGGCGCTGGG-3' Tb5motif1NdeI-Fw 5’-GGAGATATACATATGGGGCCCATGGAAGAC-3’ Tb5motif3BamHI-Rv 5’-CGCGGATCCTCACTGCAGACGCTCCTG-3’ underbar shows NdeI and BamHI site, respectively Tb5M1inv-Fw 5'-TGAGGATCCAGATCTAAGCTTGGTACC-3' Tb5M1inv-Rv 5'-TTGATGGGCAGCAAAGTGCTG-3' Tb5M3inv-Fw 5'-GACCCCAGCGCCGACGTTGAG-3' Tb5M3inv-Rv 5'-CATATGGGAGCCCTGGAAATACAGGTT-3' Tb5M2short inv-Fw 5'-CACCAGGCGGAGTGGGGACAG-3' Tb5M2short inv-Rv 5'-GCACCTCAACATCCTTGTAATCCTG-3' Tb14domain-Fw 5'-AAATTTATACATATGTCTGAACGTGAGAAACGTGTT-3' Tb14domain-Rv 5'-TATAAATTTGGATCCTCATCCAACTTTGGTGAACGC-3' Tb5E321A-Fw 5'-CATACGCACAAATGCAGGCCATG-3' Tb5E321A-Rv 5'-CCTGTGCCCACTGCTCAACGTC-3' Tb5E321H-Fw 5'-ACTACGCACAAATGCAGGCCATG-3' Tb5E321H-Rv 5'-GCTGTGCCCACTGCTCAACGTC-3' 5 Supplementary Table 2 Data collection, structure determination and refinement statistics for TbPex14p. TbPex14p Data collection * Space group P6122 Cell dimensions a, b, c (Å) 53.6, 53.6 342.8 (°) 90.0, 90.0, 120.0 Wavelength (Å) 1.000 27.57 - 1.65 (1.74 - Resolution (Å) 1.65) Rsym 0.061 (0.478) I / I 24.3 (6.2) Completeness (%) 100.0 (100.0) Redundancy 17.4 (18.3) Refinement Resolution (Å) 27.57 - 1.65 No. reflections 37018 Rwork / Rfree 0.20 /0.23 No. atoms Protein 1951 Ligand (Sulfate) 15 Water 195 B-factors Protein 33 Ligand (Sulfate) 39 Water 40 R.m.s. deviations Bond length (Å) 0.006 Bond angles (°) 0.899 a Highest-resolution shell is shown in parenthesis. Rmerge = ∑hkl∑i |Ii(hkl) - <I(hkl)>| / ∑hkl∑i Ii(hkl). c Rcryst and Rfree = ∑ ||Fobs| - |Fcalc|| / ∑|Fobs|; Rfree = R-factor for a selected subset (5%) of reflections that was not included in prior refinement calculations. d r.m.s. deviations for bond angles and lengths in regard to Engh and Huber parameters [1] b 6 1. Engh, R.A. and Huber, R. (1991) Accurate bond and angle parameters for X-ray protein structure refinement. Acta Crystallogr. A 47, 392-400. 7