The Assumptions of ANOVA
advertisement

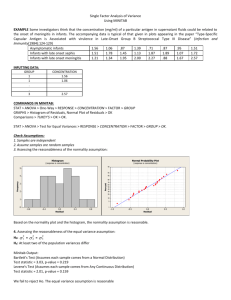
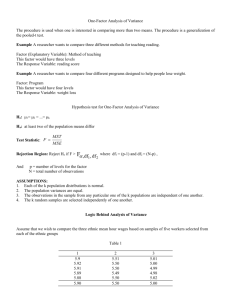
The Assumptions of ANOVA Dennis Monday Gary Klein Sunmi Lee May 10, 2005 Major Assumptions of Analysis of Variance • The Assumptions – Independence – Normally distributed – Homogeneity of variances • Our Purpose – Examine these assumptions – Provide various tests for these assumptions • Theory • Sample SAS code (SAS, Version 8.2) – Consequences when these assumptions are not met – Remedial measures Normality • Why normal? – ANOVA is an Analysis of Variance – Analysis of two variances, more specifically, the ratio of two variances – Statistical inference is based on the F distribution which is given by the ratio of two chi-squared distributions – No surprise that each variance in the ANOVA ratio come from a parent normal distribution • Calculations can always be derived no matter what the distribution is. Calculations are algebraic properties separating sums of squares. Normality is only needed for statistical inference. Normality Tests • Wide variety of tests we can perform to test if the data follows a normal distribution. • Mardia (1980) provides an extensive list for both the univariate and multivariate cases, categorizing them into two types – Properties of normal distribution, more specifically, the first four moments of the normal distribution • Shapiro-Wilk’s W (compares the ratio of the standard deviation to the variance multiplied by a constant to one) – Goodness-of-fit tests, • Kolmogorov-Smirnov D • Cramer-von Mises W2 • Anderson-Darling A2 Normality Tests proc univariate data=temp normal plot; var expvar; run; proc univariate data=temp normal plot; var normvar; run; Tests for Normality Tests for Normality Test --Statistic--- -----p Value------ Test --Statistic--- -----p Value------ Shapiro-Wilk Kolmogorov-Smirnov Cramer-von Mises Anderson-Darling W D W-Sq A-Sq Pr Pr Pr Pr Shapiro-Wilk Kolmogorov-Smirnov Cramer-von Mises Anderson-Darling W D W-Sq A-Sq Pr Pr Pr Pr 0.731203 0.206069 1.391667 7.797847 < > > > W D W-Sq A-Sq <0.0001 <0.0100 <0.0050 <0.0050 0.989846 0.057951 0.03225 0.224264 < > > > W D W-Sq A-Sq 0.6521 >0.1500 >0.2500 >0.2500 Normal Probability Plot 8.25+ | * | | | * | | * | + 4.25+ ** | ++++ ** +++ | *+++ | +++* | ++**** | ++++ ** | ++++***** | ++****** 0.25+* * ****************** +----+----+----+----+----+----+----+----+----+----+ Stem 8 7 7 6 6 5 5 4 4 3 3 2 2 1 1 0 0 Leaf 0 # 1 Boxplot * 1 1 * 2 5 4 588 3 59 00112234 56688 00011122223444 55555566667777778999999 000011111111111112222222233333334444444 ----+----+----+----+----+----+----+---- 1 1 1 3 1 2 8 5 14 23 39 * 0 0 0 0 | | | +--+--+ *-----* +-----+ Normal Probability Plot 2.3+ ++ * | ++* | +** | +** | **** | *** | **+ | ** | *** | **+ | *** 0.1+ *** | ** | *** | *** | ** | +*** | +** | +** | **** | ++ | +* -2.1+*++ +----+----+----+----+----+----+----+----+----+----+ -2 -1 0 +1 +2 Stem 22 20 18 16 14 12 10 8 6 4 2 0 -0 -2 -4 -6 -8 -10 -12 -14 -16 -18 -20 Leaf 1 7 90 047 6779 469002 2368 005546 228880077 5233446 3458447 366904459 52871 884318651 98619 60 98557220 963 584 853 0 4 8 ----+----+----+----+ Multiply Stem.Leaf by 10**-1 # 1 1 2 3 4 6 4 6 9 7 7 9 5 9 5 2 8 3 3 3 1 1 1 Boxplot | | | | | | | +-----+ | | | | *-----* | + | | | | | +-----+ | | | | | | | | Consequences of Non-Normality • F-test is very robust against non-normal data, especially in a fixed-effects model • Large sample size will approximate normality by Central Limit Theorem (recommended sample size > 50) • Simulations have shown unequal sample sizes between treatment groups magnify any departure from normality • A large deviation from normality leads to hypothesis test conclusions that are too liberal and a decrease in power and efficiency Remedial Measures for NonNormality • Data transformation • Be aware - transformations may lead to a fundamental change in the relationship between the dependent and the independent variable and is not always recommended. • Don’t use the standard F-test. – Modified F-tests • Adjust the degrees of freedom • Rank F-test (capitalizes the F-tests robustness) – Randomization test on the F-ratio – Other non-parametric test if distribution is unknown – Make up our own test using a likelihood ratio if distribution is known Independence • Independent observations – No correlation between error terms – No correlation between independent variables and error • Positively correlated data inflates standard error – The estimation of the treatment means are more accurate than the standard error shows. Independence Tests • If we have some notion of how the data was collected, we can check if there exists any autocorrelation. • The Durbin-Watson statistic looks at the correlation of each value and the value before it – Data must be sorted in correct order for meaningful results – For example, samples collected at the same time would be ordered by time if we suspect results could depend on time Independence Tests proc glm data=temp; class trt; model y = trt / p; output out=out_ds r=resid_var; run; quit; proc glm data=temp; class trt; model y = trt / p; output out=out_ds r=resid_var; run; quit; data out_ds; set out_ds; time = _n_; run; proc gplot data=out_ds; plot resid_var * time; run; quit; data out_ds; set out_ds; time = _n_; run; proc gplot data=out_ds; plot resid_var * time; run; quit; First Order Autocorrelation 0.90931 Durbin-Watson D 0.12405 First Order Autocorrelation 0.00479029 Durbin-Watson D 1.96904290 Remedial Measures for Dependent Data • First defense against dependent data is proper study design and randomization – Designs could be implemented that takes correlation into account, e.g., crossover design • Look for environmental factors unaccounted for – Add covariates to the model if they are causing correlation, e.g., quantified learning curves • If no underlying factors can be found attributed to the autocorrelation – Use a different model, e.g., random effects model – Transform the independent variables using the correlation coefficient Homogeneity of Variances • Eisenhart (1947) describes the problem of unequal variances as follows – the ANOVA model is based on the proportion of the mean squares of the factors and the residual mean squares – The residual mean square is the unbiased estimator of 2, the variance of a single observation – The between treatment mean squares takes into account not only the differences between observations, 2, just like the residual mean squares, but also the variance between treatments – If there was non-constant variance among treatments, we can replace the residual mean square with some overall variance, a2, and a treatment variance, t2, which is some weighted version of a2 – The “neatness” of ANOVA is lost Homogeneity of Variances • The omnibus (overall) F-test is very robust against heterogeneity of variances, especially with fixed effects and equal sample sizes. • Tests for treatment differences like t-tests and contrasts are severely affected, resulting in inferences that may be too liberal or conservative. Tests for Homogeneity of Variances – Levene’s Test • computes a one-way-anova on the absolute value (or sometimes the square) of the residuals, |yij – ŷi| with t-1, N – t degrees of freedom • Considered robust to departures of normality, but too conservative – Brown-Forsythe Test • a slight modification of Levene’s test, where the median is substituted for the mean (Kuehl (2000) refers to it as the Levene (med) Test) – The Fmax Test • Proportion of the largest variance of the treatment groups to the smallest and compares it to a critical value table • Tabachnik and Fidell (2001) use the Fmax ratio more as a rule of thumb rather than using a table of critical values. – Fmax ratio is no greater than 10 – Sample sizes of groups are approximately equal (ratio of smallest to largest is no greater than 4) • No matter how the Fmax test is used, normality must be assumed. Tests for Homogeneity of Variances proc glm class model means run; quit; data=temp; trt; y = trt; trt / hovtest=levene hovtest=bf; Homogeneous Variances The GLM Procedure proc glm class model means run; quit; data=temp; trt; y = trt; trt / hovtest=levene hovtest=bf; Heterogenous Variances The GLM Procedure Levene's Test for Homogeneity of Y Variance ANOVA of Squared Deviations from Group Means Source DF Sum of Squares Mean Square TRT Error 1 98 10.2533 1663.5 10.2533 16.9747 Levene's Test for Homogeneity of y Variance ANOVA of Squared Deviations from Group Means F Value Pr > F Source DF Sum of Squares Mean Square 0.60 0.4389 trt Error 1 98 10459.1 27921.5 10459.1 284.9 Brown and Forsythe's Test for Homogeneity of Y Variance ANOVA of Absolute Deviations from Group Medians Source DF Sum of Squares Mean Square TRT Error 1 98 0.7087 124.6 0.7087 1.2710 F Value Pr > F 36.71 <.0001 Brown and Forsythe's Test for Homogeneity of y Variance ANOVA of Absolute Deviations from Group Medians F Value Pr > F Source DF Sum of Squares Mean Square 0.56 0.4570 trt Error 1 98 318.3 333.8 318.3 3.4065 F Value Pr > F 93.45 <.0001 Tests for Homogeneity of Variances • SAS (as far as I know) does not have a procedure to obtain Fmax (but easy to calculate) • More importantly: VARIANCE TESTS ARE ONLY FOR ONE-WAY ANOVA WARNING: Homogeneity of variance testing and Welch's ANOVA are only available for unweighted one-way models. Tests for Homogeneity of Variances (Randomized Complete Block Design and/or Factorial Design) • In a CRD, the variance of each treatment group is checked for homogeneity • In factorial/RCBD, each cell’s variance should be checked H0: σij2 = σi’j’2, For all i,j where i ≠ i’, j ≠ j’ Tests for Homogeneity of Variances (Randomized Complete Block Design and/or Factorial Design) • • Approach 1 – – Code each row/column to its own group Run HOVTESTS as before Approach 2 – – – data newgroup; set oldgroup; if block = 1 and treat = 1 then newgroup if block = 1 and treat = 2 then newgroup if block = 2 and treat = 1 then newgroup if block = 2 and treat = 2 then newgroup if block = 3 and treat = 1 then newgroup if block = 3 and treat = 2 then newgroup Recall Levene’s Test and BrownForsythe Test are ANOVAs based on residuals Find residual for each observation Run ANOVA proc sort data=oldgroup; by treat block; run; = 1; = 2; = 3; = 4; = 5; = 6; run; proc glm data=newgroup; class newgroup; model y = newgroup; means newgroup / hovtest=levene hovtest=bf; run; quit; proc means data=oldgroup noprint; by treat block; var y; output out=stats mean=mean median=median; run; data newgroup; merge oldgroup stats; by treat block; resid = abs(mean - y); if block = 1 and treat = 1 then newgroup = 1; ……… run; proc glm data=newgroup; class newgroup; model resid = newgroup; run; quit; Tests for Homogeneity of Variances (Repeated-Measures Design) • Recall the repeated-measures set-up: Treatment a1 a2 a3 s1 s1 s1 s2 s2 s2 s3 s3 s3 s4 s4 s4 Tests for Homogeneity of Variances (Repeated-Measures Design) • As there is only one score per cell, the variance of each cell cannot be computed. Instead, four assumptions need to be tested/satisfied – Compound Symmetry • Homogeneity of variance in each column – σa12 = σa22 = σa32 • Homogeneity of covariance between columns – σa1a2 = σa2a3 = σa3a1 – No A x S Interaction (Additivity) – Sphericity • Variance of difference scores between pairs are equal – σYa1-Ya2 = σYa1-Ya3 = σYa2-Ya3 Tests for Homogeneity of Variances (Repeated-Measures Design) • Usually, testing sphericity will suffice • Sphericity can be tested using the Mauchly test in SAS proc glm data=temp; class sub; model a1 a2 a3 = sub / nouni; repeated as 3 (1 2 3) polynomial / summary printe; run; quit; Sphericity Tests Variables Transformed Variates Orthogonal Components DF 2 2 Mauchly's Criterion Det = 0 Det = 0 Chi-Square 6.01 6.03 Pr > ChiSq .056 .062 Tests for Homogeneity of Variances (Latin-Squares/Split-Plot Design) • If there is only one score per cell, homogeneity of variances needs to be shown for the marginals of each column and each row – Each factor for a latin-square – Whole plots and subplots for split-plot • If there are repititions, homogeneity is to be shown within each cell like RCBD • If there are repeated-measures, follow guidelines for sphericity, compound symmetry and additivity as well Remedial Measures for Heterogeneous Variances • Studies that do not involve repeated measures – If normality is not violated, a weighted ANOVA is suggested (e.g., Welch’s ANOVA) – If normality is violated, the data transformation necessary to normalize data will usually stabilize variances as well – If variances are still not homogeneous, non-ANOVA tests might be your option • Studies with repeated measures – For violations of sphericity • modify the degrees of freedom have been suggested. – Greenhouse-Geisser – Huynh and Feldt • Only do specific comparisons (sphericity does not apply since only two groups – sphericity implies more than two) • MANOVA • Use an MLE procedure to specify variance-covariance matrix Other Concerns • Outliers and influential points – Data should always be checked for influential points that might bias statistical inference • Use scatterplots of residuals • Statistical tests using regression to detect outliers – DFBETAS – Cook’s D References • Casella, G. and Berger, R. (2002). Statistical Inference. United States: Duxbury. • Cochran, W. G. (1947). Some Consequences When the Assumptions for the Analysis of Variances are not Satisfied. Biometrics. Vol. 3, 22-38. • Eisenhart, C. (1947). The Assumptions Underlying the Analysis of Variance. Biometrics. Vol. 3, 1-21. • Ito, P. K. (1980). Robustness of ANOVA and MANOVA Test Procedures. Handbook of Statistics 1: Analysis of Variance (P. R. Krishnaiah, ed.), 199-236. Amsterdam: NorthHolland. • Kaskey, G., et al. (1980). Transformations to Normality. Handbook of Statistics 1: Analysis of Variance (P. R. Krishnaiah, ed.), 321-341. Amsterdam: North-Holland. • Kuehl, R. (2000). Design of Experiments: Statistical Principles of Research Design and Analysis, 2nd edition. United States: Duxbury. • Kutner, M. H., et al. (2005). Applied Linear Statistical Models, 5th edition. New York: McGraw-Hill. • Mardia, K. V. (1980). Tests of Univariate and Multivariate Normality. Handbook of Statistics 1: Analysis of Variance (P. R. Krishnaiah, ed.), 279-320. Amsterdam: North-Holland. • Tabachnik, B. and Fidell, L. (2001). Computer-Assisted Research Design and Analysis. Boston: Allyn & Bacon.