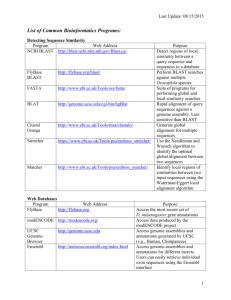
ncbi blast
advertisement

Web Databases for Drosophila An introduction to web tools, databases and NCBI BLAST Wilson Leung 01/2016 Agenda • GEP annotation project overview • Web databases for Drosophila annotation – UCSC Genome Browser – NCBI / BLAST – FlyBase – Gene Record Finder AAACAACAATCATAAATAGAGGAAGTTTTCGGAATATACGATAAGTGAAATATCGTTCT TAAAAAAGAGCAAGAACAGTTTAACCATTGAAAACAAGATTATTCCAATAGCCGTAAGA GTTCATTTAATGACAATGACGATGGCGGCAAAGTCGATGAAGGACTAGTCGGAACTGGA AATAGGAATGCGCCAAAAGCTAGTGCAGCTAAACATCAATTGAAACAAGTTTGTACATC GATGCGCGGAGGCGCTTTTCTCTCAGGATGGCTGGGGATGCCAGCACGTTAATCAGGAT ACCAATTGAGGAGGTGCCCCAGCTCACCTAGAGCCGGCCAATAAGGACCCATCGGGGGG GCCGCTTATGTGGAAGCCAAACATTAAACCATAGGCAACCGATTTGTGGGAATCGAATT TAAGAAACGGCGGTCAGCCACCCGCTCAACAAGTGCCAAAGCCATCTTGGGGGCATACG CCTTCATCAAATTTGGGCGGAACTTGGGGCGAGGACGATGATGGCGCCGATAGCACCAG CGTTTGGACGGGTCAGTCATTCCACATATGCACAACGTCTGGTGTTGCAGTCGGTGCCA TAGCGCCTGGCCGTTGGCGCCGCTGCTGGTCCCTAATGGGGACAGGCTGTTGCTGTTGG TGTTGGAGTCGGAGTTGCCTTAAACTCGACTGGAAATAACAATGCGCCGGCAACAGGAG CCCTGCCTGCCGTGGCTCGTCCGAAATGTGGGGACATCATCCTCAGATTGCTCACAATC ATCGGCCGGAATGNTAANGAATTAATCAAATTTTGGCGGACATAATGNGCAGATTCAGA ACGTATTAACAAAATGGTCGGCCCCGTTGTTAGTGCAACAGGGTCAAATATCGCAAGCT CAAATATTGGCCCAAGCGGTGTTGGTTCCGTATCCGGTAATGTCGGGGCACAATGGGGA GCCACACAGGCCGCGTTGGGGCCCCAAGGTATTTCCAAGCAAATCACTGGATGGGAGGA ACCACAATCAGATTCAGAATATTAACAAAATGGTCGGCCCCGTTGTTATGGATAAAAAA TTTGTGTCTTCGTACGGAGATTATGTTGTTAATCAATTTTATTAAGATATTTAAATAAA TATGTGTACCTTTCACGAGAAATTTGCTTACCTTTTCGACACACACACTTATACAGACA GGTAATAATTACCTTTTGAGCAATTCGATTTTCATAAAATATACCTAAATCGCATCGTC Start codon Coding region Stop codon Intron donor Intron acceptor UTR Annotation – adding labels to a sequence • • • • • • Genes: Novel or known genes, pseudogenes Regulatory Elements: Promoters, enhancers, silencers Non-coding RNA: tRNAs, miRNAs, siRNAs, snoRNAs Repeats: Transposable elements, simple repeats Structural: Origins of replication Experimental Results: – DNase I Hypersensitive sites – ChIP-chip and ChIP-Seq datasets (e.g. modENCODE) GEP Drosophila annotation projects D. melanogaster D. simulans D. sechellia D. yakuba D. erecta D. ficusphila D. eugracilis D. biarmipes D. takahashii D. elegans D. rhopaloa D. kikkawai D. bipectinata D. ananassae D. pseudoobscura D. persimilis D. willistoni D. mojavensis D. virilis D. grimshawi Reference Published Species in the Four Genomes Paper Annotation projects for Fall 2015 / Spring 2016 Manuscript in progress New species sequenced by modENCODE Phylogenetic tree produced by Thom Kaufman as part of the modENCODE project Gene annotation workflow Visualize a genomic region with evidence tracks GEP UCSC Genome Browser Identify interesting features and putative orthologs NCBI BLAST Learn about the putative D. melanogaster ortholog NCBI / FlyBase Understand the gene and isoform structure Gene Record Finder UCSC Genome Browser • Provide graphical view of genomic regions – Sequence conservation – Gene and splice site predictions – RNA-Seq and splice junction predictions • BLAT – BLAST-Like Alignment Tool – Map protein or nucleotide sequences against an assembly – Faster but less sensitive than BLAST • Table Browser – Access data used to create the graphical browser UCSC Genome Browser overview Genomic sequence Gene predictions RNA-seq Repeats Comparative genomics Evidence tracks BLASTX alignments Control how evidence tracks are displayed on the Genome Browser • Most evidence tracks have five display modes: – Hide: track is hidden – Dense: all features (including overlapping features) are displayed on a single line – Squish: overlapping features are drawn on separate lines, features are half the height compared to full mode – Pack: overlapping features are drawn on separate lines, features are the same height as full mode – Full: Each feature is displayed on its own line • Some annotation tracks (e.g. RepeatMasker) only have a subset of these display modes Two different versions of the UCSC Genome Browser Official UCSC Version http://genome.ucsc.edu Published data, lots of species, whole genomes, used for “Chimp Chunks” GEP Version http://gander.wustl.edu GEP data, parts of genomes, used for annotation of Drosophila species Additional training resources • Training section on the UCSC web site – http://genome.ucsc.edu/training/index.html – User guides – Mailing lists • OpenHelix tutorials and training materials – http://www.openhelix.com/ucsc – Pre-recorded tutorial – Reference cards UCSC GENOME BROWSER DEMO Use BLAST to detect sequence similarity • BLAST = Basic Local Alignment Search Tool • Why is BLAST popular? – Provide statistical significance for each match – Good balance of sensitivity and speed • Find local regions of similarity irrespective of where they are in the sequence Common types of BLAST programs • Except for BLASTN, all alignments are based on comparisons of protein sequences • Decide which BLAST program to use based on the type of query and subject sequences: Program BLASTN BLASTP Query Nucleotide Protein Database (Subject) Nucleotide Protein BLASTX TBLASTN TBLASTX Nucleotide → Protein Protein Nucleotide → Protein Protein Nucleotide → Protein Nucleotide → Protein Common BLAST programs use cases • BLASTN: Search for similar nucleotide sequences – Map contigs to genome, mRNAs/ESTs to genome • • • • BLASTP: Search for proteins similar to predicted genes BLASTX: Map protein / exons against genomic sequence TBLASTN: Map protein against genomic assemblies TBLASTX: Identify genes in unannotated sequences • See the BLAST Homepage and Selected Search Pages document for details: • ftp://ftp.ncbi.nlm.nih.gov/pub/factsheets/HowTo_BLASTGuide.pdf NCBI BLAST nucleotide databases • GenBank Non-Redundant Nucleotide Database (nr/nt) – Most comprehensive but some entries are low quality – Exclude sequences from whole genome assembly, ESTs • RefSeq RNA Database – mRNA and non-coding RNA entries from the NCBI Reference Sequence Project – Include real and computationally predicted sequences • Expressed Sequence Tag (EST) Database – ESTs are short single reads of cDNA clones – High error rate but useful for identifying transcribed loci NCBI BLAST protein databases • GenBank Non-Redundant Protein Database (nr) – Most comprehensive but some entries are low quality – Include sequences from both RefSeq and UniProtKB • RefSeq Protein Database – Sequences from the NCBI Reference Sequence Project – Higher quality than the nr database – Include real and computationally predicted sequences • UniProtKB / Swiss-Prot Protein Database – Manually curated proteins from literature – Real proteins with known functions – Much smaller database than either RefSeq or nr Where can I run BLAST? • NCBI BLAST web service – http://blast.ncbi.nlm.nih.gov/Blast.cgi • EBI BLAST web service – http://www.ebi.ac.uk/Tools/sss/ • FlyBase BLAST (Drosophila and other insects) – http://flybase.org/blast/ NCBI BLAST DEMO National Center for Biotechnology Information (NCBI) http://www.ncbi.nlm.nih.gov Key features of NCBI • Strengths – Most comprehensive among publicly available databases – PubMed for literature searches – Comprehensive BLAST web service • Weaknesses – Web site is large and complex – Quality of GenBank records may vary • Use cases – Perform BLAST searches against Refseq, nr/nt databases – Compare one sequence against another (bl2seq) FlyBase - Database for the Drosophila research community http://flybase.org/ Key Features of FlyBase • Lots of ancillary data for each gene in Drosophila • Curation of literature for each gene • Reference Drosophila annotations for all the other databases (including NCBI) • Fast release cycle (6-8 releases per year) • Use cases – Species-specific BLAST searches – Genome browser (GBrowse) and access datasets for 20 Drosophila species Web databases and tools • Many genome databases available – Be aware of different annotation releases – Use FlyBase as the canonical reference • Web databases are being updated constantly – Update GEP materials before semester starts – Discrepancies in exercise screenshots – Minor changes in search results – Let us know about errors or revisions FLYBASE DEMO Gene Record Finder http://gander.wustl.edu/~wilson/dmelgenerecord/index.html Key features of the Gene Record Finder • List of unique coding and non-coding exons for each gene in D. melanogaster • CDS and exon usage maps for each isoform • Optimized for exon-by-exon annotation strategy • Slower update release cycle than FlyBase – Database is updated every semester • Use cases: – Get amino acid sequences and nucleotide sequences of each exon for BLAST 2 Sequences (bl2seq) searches GENE RECORD FINDER DEMO Summary • GEP annotation project seeks to generate high quality manually curated gene models for multiple Drosophila species • Use BLAST to characterize a genomic sequence • Use web databases to gather information on a gene – – – – UCSC Genome Browser NCBI FlyBase Gene Record Finder Questions? http://www.flickr.com/photos/jac_opo/240254763/sizes/l/ Brief overview of the BLAST algorithm • BLAST is based on the Smith-Waterman algorithm but use the following heuristics: – Build word list with the query and subject sequences • Word size = 3 for protein, 11 for nucleotide – Generate a list of high-scoring words • Determine by scoring system or scoring matrix – Scan database for exact matches to high-scoring words – Extend matches to generate High-scoring Segment Pairs – Merge multiple HSP’s into a longer alignment – Calculate E-value for the alignments – Report alignments below E-value threshold Ensembl Metazoa: Databases for 12 Drosophila species http://metazoa.ensembl.org/index.html Key features of Ensembl Metazoa • Lots of ancillary data for each gene • Data for 12 Drosophila species available • Detailed information on each gene available at the transcript, peptide, and exon level • Not always up-to-date – Annotations are from FlyBase Release 6.02 • Use cases – Get amino acid sequences and nucleotide sequences of each exon for bl2seq searches – Perform species-specific BLAST searches