Post-transcriptional Gene Silencing (PTGS)
advertisement
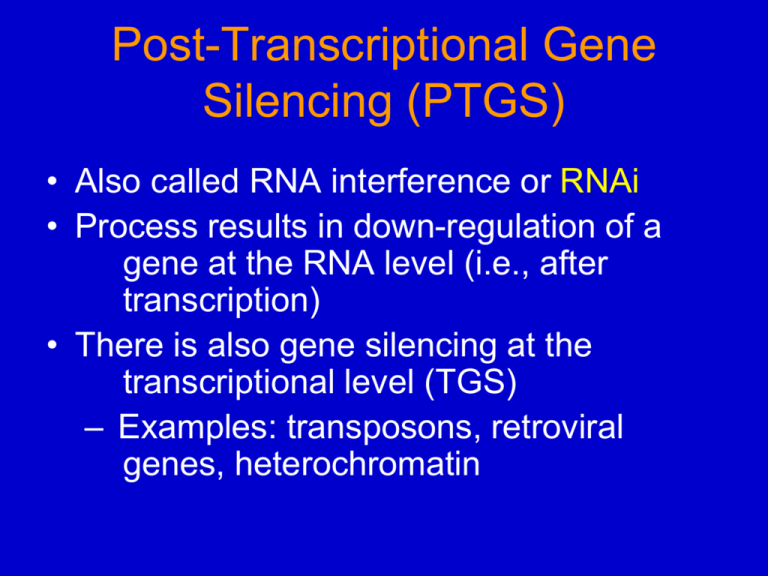
Post-Transcriptional Gene Silencing (PTGS) • Also called RNA interference or RNAi • Process results in down-regulation of a gene at the RNA level (i.e., after transcription) • There is also gene silencing at the transcriptional level (TGS) – Examples: transposons, retroviral genes, heterochromatin • PTGS is heritable, although it can be modified in subsequent cell divisions or generations – Ergo, it is an epigenetic phenomenon Epigenetics - refers to heritable changes in phenotype or gene expression caused by mechanisms other than changes in the underlying DNA sequence. Antisense Technology • Used from ~1980 on, to repress specific genes – Alternative to gene knock-outs, which were/are very difficult to do in higher plants and animals • Theory: by introducing an antisense gene (or asRNA) into cells, the asRNA would “zip up” the complementary mRNA into a dsRNA that would not be translated • The “antisense effect” was highly variable, and in light of the discovery of RNAi, asRNA probably inhibited its target by inducing RNAi rather than inhibiting translation. Discovery of PTGS • First discovered in plants – (R. Jorgensen, 1990) • When Jorgensen introduced a re-engineered gene into petunia that had a lot of homology with an endogenous petunia gene, both genes became suppressed! – Also called Co-suppression – Suppression was mostly due to increased degradation of the mRNAs (from the endogenous and introduced genes) Discovery of PTGS (cont.) • Involved attempts to manipulate pigment synthesis genes in petunia • Genes were enzymes of the flavonoid/ anthocyanin pathway: – CHS: chalcone synthase – DFR: dihydroflavonol reductase • When these genes were introduced into petunia using a strong viral promoter, mRNA levels dropped and so did pigment levels in many transgenics. Flavonoid/anthocyanin pathway in plants Strongly pigmented compounds DFR construct introduced into petunia CaMV - 35S promoter from Cauliflower Mosaic Virus DFR cDNA – cDNA copy of the DFR mRNA (intronless DFR gene) T Nos - 3’ processing signal from the Nopaline synthase gene Flowers from 3 different transgenic petunia plants carrying copies of the chimeric DFR gene above. The flowers had low DFR mRNA levels in the non-pigmented areas, but gene was still being transcribed. • RNAi discovered in C. elegans (first animal) while attempting to use antisense RNA in vivo Craig Mello Andrew Fire (2006 Nobel Prize in Physiology & Medicine) – Control “sense” RNAs also produced suppression of target gene! – sense RNAs were contaminated with dsRNA. – dsRNA was the suppressing agent. Double-stranded RNA (dsRNA) induced interference of the Mex-3 mRNA in the nematode C. elegans. Antisense RNA (c) or dsRNA (d) for the mex3 (mRNA) was injected into C. elegans ovaries, and then mex3 mRNA was detected in embryos by in situ hybridization with a mex-3 probe. (a) control embryo (b) control embryo hyb. with mex-3 probe Conclusions: (1) dsRNA reduced mex-3 mRNA better than antisense mRNA. (2) the suppressing signal moved from cell to cell. Fig. 16.29 PTGS (RNAi) occurs in wide variety of Eukaryotes: – Angiosperms – C. elegans (nematode) – Drosophila – Mammalian cells – Chlamydomonas (unicellular – Neurospora, but not in Yeast! Mechanism of RNAi: Role of Dicer 1. 2. 3. Cells (plants and animals) undergoing RNAi contained small fragments (~25 nt) of the RNA being suppressed. A nuclease (Dicer) was purified from Drosophila embryos that still had small RNA fragments associated with it, both sense and antisense. The Dicer gene is found in all organisms that exhibit RNAi, and mutating it inhibits the RNAi effect. Conclusion: Dicer is the endonuclease that degrades dsRNA into 21-24 nt fragments, and in higher eukaryotes also pulls the strands apart via intrinsic helicase activity. Generation of 21-23 nt fragments of target RNA in a RNAi-competent Droso. embryo lysate/extract. 32P-labeled ds luciferase (luc) RNAs, either Pp or Rr, were added to reactions 2-10 in the presence or absence of the corresponding mRNA. The dsRNAs were labeled on the sense (s), antisense (a) or both (a/s) strands. Lanes 11, 12 contained 32P-labeled, capped, antisense Rr-luc RNA. Fig. 16.30 The dsRNA that is added dictates where the destabilized mRNA is cleaved. The dsRNAs A, B, or C were added to the Drosophila extract together with a Rr-luc mRNA that is 32P-labeled at the 5’ end. The RNA was then analyzed on a polyacrylamide gel and autoradiographed. Results: the products of Rr-luc mRNA degradation triggered by dsRNA B are ~100nt longer than those triggered by dsRNA C (and ~100 nt longer again for dsRNA A-induced degradation). Fig. 16.31 Model for RNAi By “Dicer” 21-23 nt RNAs ATP-dependent Helicase or Dicer Very efficient process because many small interfering RNAs (siRNAs) generated from a larger dsRNA. Fig. 16.39, 3rd Ed. Active siRNA complexes = RISC - contain Argonaute instead of Dicer In plants, fungi, C. elegans & Drosophila, a RNAdependent RNA polymerase (RDR) is involved in the initiation (b) or amplification (c) of silencing (RNAi). CBP and PABP block access for RDR. PABP missing. D. Baulcombe 2004 Nature 431:356 Why RNAi silencing? • Most widely held view is that RNAi evolved to protect the genome from viruses (and perhaps transposons or mobile DNAs). • Some viruses have proteins that suppress silencing: 1. HCPro - first one identified, found in plant potyviruses (V. Vance) 2. P19 - tomato bushy stunt virus, binds to siRNAs and prevents RISC formation (D. Baulcombe). 3. Tat - RNA-binding protein from HIV Micro RNAs (MiRNAs) • Recently, very small (micro) MiRNAs have been discovered in plants and animals. • They resemble siRNAs, and they regulate specific mRNAs by promoting their degradation or repressing their translation. • New use for the RNAi mechanism besides defense. Comparison of Mechanisms of MiRNA Biogenesis and Action DCL1 mutant Better complementarity of MiRNAs and targets in plants. Summary of differences between plant and animal MiRNA systems Plants Animals # of miRNA genes: 100-200 100-500 Location in genome: intergenic regions Intergenic regions, introns Clusters of miRNAs: Uncommon Common MiRNA biosynthesis: Dicer-like Drosha, Dicer Mechanism of repression mRNA cleavage Translational repression Location of miRNA target in a gene: Predominantly the open-reading frame Predominantly the 3′-UTR Generally one Generally multiple Regulatory genes crucial for development, enzymes Regulatory genes—crucial for development, structural proteins, enzymes # of miRNA binding sites in a target gene: Functions of known target genes: