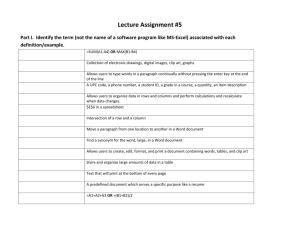
Microsoft Research
Faculty Summit 2007
Colonies Of
Synchronizing Agents:
Molecules, Cells,
And Tissues
Matteo Cavaliere – MSR – UNITN CoSBi
(Trento, Italy)
Giuditta Franco - University of Verona, Italy
Natasha Jonoska – University of South Florida
Sean Sedwards – MSR – UNITN CoSBi
(Trento, Italy)
Motivation
Model
intuitiveness,
transparency, scalability,
composability,
expressivity, simplicity,
analysability …
Reality
Analysis
Interpretation
Understanding
and Prediction
Formalization…
Petri nets process algebra
ODE statistical mechanics
rewriting
automata
Efficient simulation…
Analytical solution…
Role Of Computer Science
Intuition
Experiments
Intuition
Mathematical
model
CS
The problem:
Human intuition is the limiting step
Computational
Model
Role Of Computer Science
Inference
Experiments
CS
Computational
Model
The goal:
Formalise and automate
Analysis
Mathematical
Model
A Membrane System
hierarchical system
of compartments
with membranes
multisets of floating
objects local to regions
1
a
a
0
2
b
4
a a
ab
b 3
c c
a
b+aa+c
a+bc
multisets of objects
attached to membranes
plus transport rules
c
b
ab
local ‘chemical’
rules based on
multiset rewriting
b+cb+a
system
environment
a+bc
conflicts between rules are
resolved non-deterministically
Knee Injury
The important actors:
B', C' lining cells
altered hyaluronan (HA) molecules h’
activated macrophages D’
Knee tissue in healthy state
Knee tissue after injury
Knee Injury Model
Regular cell turnover of the
system in a homeostatic state
Knee Injury Model
Gravity signals s (injury)
instigates a cascade of
biochemical interactions
(the healing process)
G. Franco, N. Jonoska, B. Osborn, A. Plaas, Knee Joint Injury
and Repair Modeled by Membrane Systems, Biosystems, to appear.
Computational Issues
Formal description and
analysis of the healing process
Confirmed structural importance
of hyaluronan for tissue repair
Analysis using techniques
from symbolic dynamics
The system is non-deterministic
Represents lack of knowledge
and innate stochasticity
Creates complexity for analysis
Potential parallelization (e.g., on a cluster)
Colonies Of Synchronizing Agents
Generalized version of Membrane Systems
Population of enclosed regions
(agents) in 3D containing objects
Internal rewriting rules (chemistry)
Pairwise synchronization rules
Synchronized rewriting (synchronized chemistry)
Passage of objects (molecules) between regions
Plus movement, division and deletion rules
Agents may represent molecules or cells
A colony may be a tissue or a solution
Colonies Of Synchronizing Agents
10
ab
baa
100
bba
ac
26
a b
cc
Number of agents of type
Initial contents of agent
Agents (cells) contain multisets of objects
(molecules) and are acted upon by rules (reactions)
chemistry
synchronization
deletion
[a,b] [c,d]
[a] [b] [c] [d]
[a] λ
movement
[a] (a,b,g)[b]
division
[a] [c] [d]
Having space, movement and division allows us to model complex spatiotemporal behaviour and structures, e.g., morphogenesis, quorum sensing…
Internal Rules
abc
ba
abc
bc
ba
b
ab
b
abc
bc
ab
b
[a,b,c,a] → [b,a]
Intracellular mechanisms, e.g., chemistry
Synchronization Rules
abc
ba
abc
bc
aa
ba
ab
b
abc
bb
ab
b
[a,b,c] [c,c] → [a,a] [c,b]
Intercellular mechanisms, e.g., signalling
Evolution Of Colonies
Global behaviour of a colony is
obtained using just internal rules +
synchronization rules
Overall behaviour is more complex than
the sum of the individual components
Robustness Of Colonies
Robust behaviour is biologically important
A robust colony
The behaviour does not change critically
if one or more agents cease to exist or if
one or more rules stop working
There are (efficient) algorithms
to check if a colony is robust*
M. Cavaliere, R. Mardare, S. Sedwards, Colonies of Synchronizing Agents: An Abstract Model of Intracellular and
Intercellular Processes, Int. Work. on Automata for Cellular and Molecular Computing, Budapest, 2007.
Why Simulate?
Modelling
power
Behavioural
complexity
Need to
simulate
maximal
…
…
minimal
Difficulty of
deciding properties
(analysability)
Simulation Complexity
Complexity of each step
of a stochastic simulation
Membrane system with M reactions: O(M)
CSA with N agents, no
synchronization: O(NM)
CSA with N agents, space and
synchronization: O(N2M)
Optimised algorithm: O(NM)
Optimised and distributed algorithm: O(NM½)
Prospects
More complex biological models
E.g., immune system, cell cycle, evolution
Model checking algorithms
Distributed implementation of CSAs
Thank You For Your Attention
Contributors:
Matteo Cavaliere – MSR – UNITN CoSBi (Trento, Italy)
Sean Sedwards – MSR – UNITN CoSBi (Trento, Italy)
Giuditta Franco - Department of Computer Science, University of Verona, Italy
Natasha Jonoska – Department of Computer Science, University of South Florida
Barbara Osborn - Department of Internal Medicine, University of South Florida
Anna Plaas - Department of Internal Medicine, University of South Florida
© 2007 Microsoft Corporation. All rights reserved. Microsoft, Windows, Windows Vista and other product names are or may be registered trademarks and/or trademarks in the U.S. and/or other
countries. The information herein is for informational purposes only and represents the current view of Microsoft Corporation as of the date of this presentation. Because Microsoft must respond to
changing market conditions, it should not be interpreted to be a commitment on the part of Microsoft, and Microsoft cannot guarantee the accuracy of any information provided after the date of
this presentation. MICROSOFT MAKES NO WARRANTIES, EXPRESS, IMPLIED OR STATUTORY, AS TO THE INFORMATION IN THIS PRESENTATION.