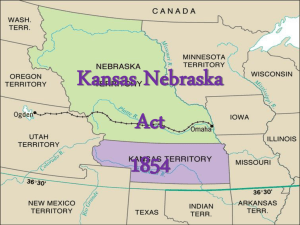
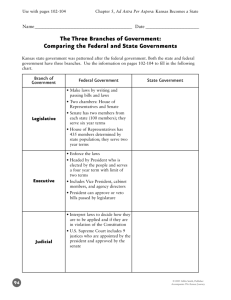
Math-20021107 - Kansas State University
advertisement

KSU Math Department Colloquium
Graphical Models of Probability
for Causal Reasoning
Thursday 07 November 2002
William H. Hsu
Laboratory for Knowledge Discovery in Databases
Department of Computing and Information Sciences
Kansas State University
http://www.kddresearch.org
This presentation is:
http://www.kddresearch.org/KSU/CIS/Math-20021107.ppt
Kansas State University KDD Lab (www.kddresearch.org)
Kansas State University
Department of Computing and Information Sciences
Overview
• Graphical Models of Probability
–
–
–
–
Markov graphs
Bayesian (belief) networks
Causal semantics
Direction-dependent separation (d-separation) property
• Learning and Reasoning: Problems, Algorithms
– Inference: exact and approximate
• Junction tree – Lauritzen and Spiegelhalter (1988)
• (Bounded) loop cutset conditioning – Horvitz and Cooper (1989)
• Variable elimination – Dechter (1996)
– Structure learning
• K2 algorithm – Cooper and Herskovits (1992)
• Variable ordering problem – Larannaga (1996), Hsu et al. (2002)
• Probabilistic Reasoning in Machine Learning, Data Mining
• Current Research and Open Problems
Kansas State University KDD Lab (www.kddresearch.org)
Kansas State University
Department of Computing and Information Sciences
Stages of Data Mining and
Knowledge Discovery in Databases
Adapted from Fayyad, Piatetsky-Shapiro, and Smyth (1996)
Kansas State University KDD Lab (www.kddresearch.org)
Kansas State University
Department of Computing and Information Sciences
Graphical Models Overview [1]:
Bayesian Networks
•
Conditional Independence
– X is conditionally independent (CI) from Y given Z (sometimes written X Y | Z) iff
P(X | Y, Z) = P(X | Z) for all values of X, Y, and Z
– Example: P(Thunder | Rain, Lightning) = P(Thunder | Lightning) T R | L
•
Bayesian (Belief) Network
– Acyclic directed graph model B = (V, E, ) representing CI assertions over
– Vertices (nodes) V: denote events (each a random variable)
– Edges (arcs, links) E: denote conditional dependencies
•
•
n
Markov Condition for BBNs (Chain Rule): P X , X , , X P X | parents X
1
2
n
i
i
i 1
Example BBN
Exposure-To-Toxins
Serum Calcium
Age X1
X3
Cancer
X6
X5
Gender X2
X4
Smoking
X7
Tumor
Lung
Descendants
NonDescendants
Parents
P(20s, Female, Low, Non-Smoker, No-Cancer, Negative, Negative)
= P(T) · P(F) · P(L | T) · P(N | T, F) · P(N | L, N) · P(N | N) · P(N | N)
Kansas State University KDD Lab (www.kddresearch.org)
Kansas State University
Department of Computing and Information Sciences
Graphical Models Overview [2]:
Markov Blankets and d-Separation Property
Motivation: The conditional independence status of
nodes within a BBN might change as the availability of
evidence E changes. Direction-dependent separation (dseparation) is a technique used to determine conditional
independence of nodes as evidence changes.
Definition: A set of evidence nodes E d-separates two
sets of nodes X and Y if every undirected path from a
node in X to a node in Y is blocked given E.
A path is blocked if one of three conditions holds:
X
From S. Russell & P. Norvig (1995)
E
(1)
Z
(2)
Z
(3)
Z
Y
Adapted from J. Schlabach (1996)
Kansas State University KDD Lab (www.kddresearch.org)
Kansas State University
Department of Computing and Information Sciences
Graphical Models Overview [3]:
Inference Problem
Multiply-connected case: exact, approximate inference are #P-complete
Adapted from slides by S. Russell, UC Berkeley
Kansas State University KDD Lab (www.kddresearch.org)
http://aima.cs.berkeley.edu/
Kansas State University
Department of Computing and Information Sciences
Other Topics in Graphical Models [1]:
Temporal Probabilistic Reasoning
•
Goal: Estimate P(X ti | y 1r )
•
Filtering: r = t
Adapted from Murphy (2001), Guo (2002)
– Intuition: infer current state from observations
– Applications: signal identification
– Variation: Viterbi algorithm
•
Prediction: r < t
– Intuition: infer future state
– Applications: prognostics
•
Smoothing: r > t
– Intuition: infer past hidden state
– Applications: signal enhancement
•
CF Tasks
– Plan recognition by smoothing
– Prediction cf. WebCANVAS – Cadez et al. (2000)
Kansas State University KDD Lab (www.kddresearch.org)
Kansas State University
Department of Computing and Information Sciences
Other Topics in Graphical Models [2]:
Learning Structure from Data
•
General-Case BBN Structure Learning: Use Inference to Compute Scores
•
Optimal Strategy: Bayesian Model Averaging
– Assumption: models h H are mutually exclusive and exhaustive
– Combine predictions of models in proportion to marginal likelihood
• Compute conditional probability of hypothesis h given observed data D
• i.e., compute expectation over unknown h for unseen cases
• Let h structure, parameters CPTs
P x m 1 | D P x 1 , x 2 , , x n | x 1 , x 2 , , x m
P x m 1 | D, h P h | D
hH
Posterior Score
Marginal Likelihood
Prior over Parameters
P h | D P D | h P h
P h P D | h, Θ P Θ | h dΘ
Prior over Structures
Kansas State University KDD Lab (www.kddresearch.org)
Likelihood
Kansas State University
Department of Computing and Information Sciences
Propagation Algorithm in Singly-Connected
Bayesian Networks – Pearl (1983)
C1
Upward (child-toparent) messages
C2
’ (Ci’) modified during
message-passing phase
C3
Downward messages
C4
C5
P’ (Ci’) is computed during
message-passing phase
C6
Multiply-connected case: exact, approximate inference are #P-complete
(counting problem is #P-complete iff decision problem is NP-complete)
Adapted from Neapolitan (1990), Guo (2000)
Kansas State University KDD Lab (www.kddresearch.org)
Kansas State University
Department of Computing and Information Sciences
Inference by Clustering [1]: Graph Operations
(Moralization, Triangulation, Maximal Cliques)
Find Maximal Cliques
A1
Clq4
Clq1
A
1
B
2
Moralize
A
Bayesian Network
(Acyclic Digraph)
A
G
E
D
8
C
G
E
D
B2
G
5
H
G5
E3
E3
G5
E3
C4
Clq3
C4
H
7
G5
C4
Clq6
F
B
Clq2
C
4
F
B
F
6
E
3
F6
B2
Triangulate
C4
D8
Clq5
H7
C
D
H
Adapted from Neapolitan (1990), Guo (2000)
Kansas State University KDD Lab (www.kddresearch.org)
Kansas State University
Department of Computing and Information Sciences
Inference by Clustering [2]:
Junction Tree – Lauritzen & Spiegelhalter (1988)
Input: list of cliques of triangulated, moralized graph Gu
Output:
Tree of cliques
Separators nodes Si,
Residual nodes Ri and potential probability (Clqi) for all cliques
Algorithm:
1. Si = Clqi (Clq1 Clq2 … Clqi-1)
2. Ri = Clqi - Si
3. If i >1 then identify a j < i such that Clqj is a parent of Clqi
4. Assign each node v to a unique clique Clqi that v c(v) Clqi
5. Compute (Clqi) = f(v) Clqi = P(v | c(v)) {1 if no v is assigned to Clqi}
6. Store Clqi , Ri , Si, and (Clqi) at each vertex in the tree of cliques
Adapted from Neapolitan (1990), Guo (2000)
Kansas State University KDD Lab (www.kddresearch.org)
Kansas State University
Department of Computing and Information Sciences
Inference by Clustering [3]:
Clique-Tree Operations
Ri: residual nodes
Si: separator nodes
(Clqi): potential probability of Clique i
A1
Clq4
Clq1
F6
B2
Clq2
B2
BEC
Clq2 = {B,E,C}
R2 = {C,E}
S2 = { B }
G5
E3
C4
Clq3
C4
G5
Clq3 = {E,C,G}
R3 = {G}
S3 = { E,C }
C4
Clq6
C4
EGF
Clq5
H7
Clq4 = {E, G, F}
R4 = {F}
S4 = { E,G }
Adapted from Neapolitan (1990), Guo (2000)
Kansas State University KDD Lab (www.kddresearch.org)
B
(Clq2) = P(C|B,E)
Clq2
ECG EC
(Clq3) = 1
Clq3
EG
(Clq4) =
P(E|F)P(G|F)P(F)
D8
(Clq1) = P(B|A)P(A)
Clq1
G5
E3
E3
AB
Clq1 = {A, B}
R1 = {A, B}
S1 = {}
CG
(Clq5) = P(H|C,G)
CGH
Clq5
Clq4
Clq5 = {C, G,H}
R5 = {H}
S5 = { C,G }
CD
Clq6 = {C, D}
R5 = {D}
S5 = { C}
(Clq2) = P(D|C)
C
Clq6
Kansas State University
Department of Computing and Information Sciences
Inference by Loop Cutset Conditioning
Age = [0, 10)
Split vertex in
undirected cycle;
condition upon each
of its state values
X1,1
Age = [10, 20)
X1,2
Exposure-ToToxins
X3
Age = [100, )
X1,10
X2
Gender
Serum Calcium
Cancer
X6
X5
X4
Smoking
•
Deciding Optimal Cutset: NP-hard
•
Current Open Problems
X7
Lung Tumor
Number of network
instantiations:
Product of arity of
nodes in minimal loop
cutset
Posterior: marginal
conditioned upon
cutset variable values
– Bounded cutset conditioning: ordering heuristics
– Finding randomized algorithms for loop cutset optimization
Kansas State University KDD Lab (www.kddresearch.org)
Kansas State University
Department of Computing and Information Sciences
Inference by Variable Elimination [1]:
Intuition
Adapted from slides by S. Russell, UC Berkeley
Kansas State University KDD Lab (www.kddresearch.org)
http://aima.cs.berkeley.edu/
Kansas State University
Department of Computing and Information Sciences
Inference by Variable Elimination [2]:
Factoring Operations
Adapted from slides by S. Russell, UC Berkeley
Kansas State University KDD Lab (www.kddresearch.org)
http://aima.cs.berkeley.edu/
Kansas State University
Department of Computing and Information Sciences
Inference by Variable Elimination [3]:
Example
P(A), P(B|A), P(C|A), P(D|B,A), P(F|B,C), P(G|F)
Season
A
Sprinkler
Rain
B
C
F
D
Wet
Manual
Watering
G
G
P(G|F) G=1
D
P(D|B,A)
F
P(F|B,C)
B
P(B|A)
C
P(C|A)
A
P(A)
Slippery
P(A|G=1) = ?
d = < A, C, B, F, D, G >
λG(f) = ΣG=1 P(G|F)
Adapted from Dechter (1996), Joehanes (2002)
Kansas State University KDD Lab (www.kddresearch.org)
Kansas State University
Department of Computing and Information Sciences
Genetic Algorithms for Parameter Tuning in
Bayesian Network Structure Learning
Dtrain (Inductive Learning)
D:
Training Data
[2] Representation Evaluator
for Learning Problems
Dval (Inference)
I e:
Inference Specification
α
f(α)
Representation
Candidate
Fitness
Representation
[1] Genetic Algorithm
Genetic Wrapper for
Change of Representation
and Inductive Bias Control
α̂
Optimized
Representation
Kansas State University KDD Lab (www.kddresearch.org)
Kansas State University
Department of Computing and Information Sciences
Supervised and Unsupervised Learning:
Decision Support in Insurance Pricing
Hsu, Welge, Redman, Clutter (2002)
Data Mining and Knowledge Discovery, 6(4):361-391
Kansas State University KDD Lab (www.kddresearch.org)
Kansas State University
Department of Computing and Information Sciences
Computational Genomics and
Microarray Gene Expression Modeling
[A] Structure
Learning
G2
D: Data (User, Microarray)
G1
G4
G3
G5
G = (V, E)
[B] Parameter
Estimation
G2
Treatment 1
(Control)
Treatment 2
(Pathogen)
Learning
Environment
Messenger RNA cDNA
(mRNA) Extract 1
G1
Dval (Model Validation by Inference)
G4
G5
G3
B = (V, E, )
Specification Fitness
(Inferential Loss)
Messenger RNA
(mRNA) Extract 2 cDNA
DNA Hybridization Microarray
(under LASER)
Adapted from Friedman et al. (2000)
http://www.cs.huji.ac.il/labs/compbio/
Kansas State University KDD Lab (www.kddresearch.org)
Kansas State University
Department of Computing and Information Sciences
DESCRIBER: An Experimental
Intelligent Filter
Users of
Scientific
Document
Repository
Example Queries:
• What experiments have found cell cycle-regulated
metabolic pathways in Saccharomyces?
• What codes and microarray data were used, and
why?
Data Entity and Source Code Repository Index for Bioinformatics Experimental Research
Personalized Interface
Historical
Use Case & Query Data
New Queries
Domain-Specific
Collaborative Filtering
Learning and Inference
Components
Decision Support
Models
Interface(s) to Distributed Repository
Domain-Specific Repositories
Experimental Data
Source Codes and Specifications
Data Models
Ontologies
Models
Kansas State University KDD Lab (www.kddresearch.org)
Kansas State University
Department of Computing and Information Sciences
Tools for Building Graphical Models
• Commercial Tools: Ergo, Netica, TETRAD, Hugin
• Bayes Net Toolbox (BNT) – Murphy (1997-present)
– Distribution page
http://http.cs.berkeley.edu/~murphyk/Bayes/bnt.html
– Development group
http://groups.yahoo.com/group/BayesNetToolbox
• Bayesian Network tools in Java (BNJ) – Hsu et al. (1999-present)
– Distribution page
http://bndev.sourceforge.net
– Development group
http://groups.yahoo.com/group/bndev
– Current (re)implementation projects for KSU KDD Lab
• Continuous state: Minka (2002) – Hsu, Guo, Perry, Boddhireddy
• Formats: XML BNIF (MSBN), Netica – Guo, Hsu
• Space-efficient DBN inference – Joehanes
• Bounded cutset conditioning – Chandak
Kansas State University KDD Lab (www.kddresearch.org)
Kansas State University
Department of Computing and Information Sciences
References [1]:
Graphical Models and Inference Algorithms
• Graphical Models
– Bayesian (Belief) Networks tutorial – Murphy (2001)
http://www.cs.berkeley.edu/~murphyk/Bayes/bayes.html
– Learning Bayesian Networks – Heckerman (1996, 1999)
http://research.microsoft.com/~heckerman
• Inference Algorithms
– Junction Tree (Join Tree, L-S, Hugin): Lauritzen & Spiegelhalter (1988)
http://citeseer.nj.nec.com/huang94inference.html
– (Bounded) Loop Cutset Conditioning: Horvitz & Cooper (1989)
http://citeseer.nj.nec.com/shachter94global.html
– Variable Elimination (Bucket Elimination, ElimBel): Dechter (1986)
http://citeseer.nj.nec.com/dechter96bucket.html
– Recommended Books
• Neapolitan (1990) – out of print; see Pearl (1988), Jensen (2001)
• Castillo, Gutierrez, Hadi (1997)
• Cowell, Dawid, Lauritzen, Spiegelhalter (1999)
– Stochastic Approximation http://citeseer.nj.nec.com/cheng00aisbn.html
Kansas State University KDD Lab (www.kddresearch.org)
Kansas State University
Department of Computing and Information Sciences
References [2]:
Machine Learning, KDD, and Bioinformatics
• Machine Learning, Data Mining, and Knowledge Discovery
– K-State KDD Lab: literature survey and resource catalog (2002)
http://www.kddresearch.org/Resources
– Bayesian Network tools in Java (BNJ): Hsu, Guo, Joehanes, Perry,
Thornton (2002)
http://bndev.sourceforge.net
– Machine Learning in Java (BNJ): Hsu, Louis, Plummer (2002)
http://mldev.sourceforge.net
– NCSA Data to Knowledge (D2K): Welge, Redman, Auvil, Tcheng, Hsu
http://www.ncsa.uiuc.edu/STI/ALG
• Bioinformatics
– European Bioinformatics Institute Tutorial: Brazma et al. (2001)
http://www.ebi.ac.uk/microarray/biology_intro.htm
– Hebrew University: Friedman, Pe’er, et al. (1999, 2000, 2002)
http://www.cs.huji.ac.il/labs/compbio/
– K-State BMI Group: literature survey and resource catalog (2002)
http://www.kddresearch.org/Groups/Bioinformatics
Kansas State University KDD Lab (www.kddresearch.org)
Kansas State University
Department of Computing and Information Sciences
Acknowledgements
• Kansas State University Lab for Knowledge Discovery in Databases
– Graduate research assistants: Haipeng Guo (hpguo@cis.ksu.edu), Roby
Joehanes (robbyjo@cis.ksu.edu)
– Other grad students: Prashanth Boddhireddy, Siddharth Chandak, Ben
B. Perry, Rengakrishnan Subramanian
– Undergraduate programmers: James W. Plummer, Julie A. Thornton
• Joint Work with
– KSU Bioinformatics and Medical Informatics (BMI) group: Sanjoy Das
(EECE), Judith L. Roe (Biology), Stephen M. Welch (Agronomy)
– KSU Microarray group: Scot Hulbert (Plant Pathology), J. Clare Nelson
(Plant Pathology), Jan Leach (Plant Pathology)
– Kansas Geological Survey, Kansas Biological Survey, KU EECS
• Other Research Partners
– NCSA Automated Learning Group (Michael Welge, Tom Redman, David
Clutter, Lisa Gatzke)
– University of Manchester (Carole Goble, Robert Stevens)
– The Institute for Genomic Research (John Quackenbush, Alex Saeed)
– International Rice Research Institute (Richard Bruskiewich)
Kansas State University KDD Lab (www.kddresearch.org)
Kansas State University
Department of Computing and Information Sciences