BNP & NTPro-BNP - Al
advertisement

Characterisation of Neuroprotective
PDZ binding peptides
by Jamie Al-Nasir
Project supervisor: Prof. Brian Austen
Presented as part of M.Pharm project at:
Kingston university and St. Georges Hospital medical school, 24/01/2011
What are PDZ domains?
◦ Recall that domains are regions of a protein with a particular sequence
◦ PDZ domains are important regions of intracellular signaling proteins
◦ They function as modular protein-protein signalling complexes in Eukaryotic cells
◦ PDZ acquires its name from the first proteins found to contain these regions
PSD-95 (Post-Synaptic-Density, 95 kDa),
DLG (Drasophila melangolaster Discs Large Protein) and
ZO-1 (Zonula occludens 1)
◦ Some proteins contain more than one PDZ region
◦ PDZ domains have been found in over 250
proteins across a wide range of organisms from
Drasophila to Humans
◦ PDZs bind to complementary sequences at C-termini
◦ Implicated in a wide range of cellular processes
◦ – therefore an extremely useful drug target
◦
PDZ domains and Neuroprotective binding
◦ PDZ Domains are found in the NMDA receptor complex where they function as
signaling scaffolds that mediate protein-protein interactions (1 and 2, figure below)
◦ In normal physiological function Glutamate is a neurotransmitter that acts on NMDA
receptors resulting in the activation of intracellular processes
◦ Excesses of Glutamate however can cause downstream toxicity, termed Excitotoxicity
- a mechanism implicated in a variety of neurologically degenerative disorders such
as Alzheimers, Parkinsons, epilepsy and stroke.
This pathological process results in
damage to the neurons!
◦ PSD-95 is a PDZ containing protein which
forms part of the NMDA receptor complex
disruption of which can mitigate and reduce
glutamate mediated neuro-degeneration.
◦ Prof. Austen and his team at St. Georges
Academic Neuroscience department are
working to develop PDZ binding ligands to
achieve this therapeutic objective.
The NMDA receptor complex
1) Glutamate binds on NR2A sub-unit
2) Ca2+ pumped into intracellular space
3) Intracellular signaling pathways
are activated
4) This is coupled by PSD-95 (pink) containing
two PDZ domains (1 and 2).
5) Normal response
OR Pathological response if prolonged
activation (in presence of excess glutamate)
Evidence favoring this approach
◦ Immunopreciptation studies by Prof. Austen et al confirm that the NR2A subunit
interacts with PMCA2b via PSD-95.
◦ Spaller, Pizerchio et al have demonstrated that disrupting this interaction using
peptides generated from PDZ binding proteins decreases the vulnerability of neurons
to excitotoxicity
Objectives
◦ “To alter the function of the NMDA receptor complex by targeting PDZ1 domain of
PSD-95 thereby disrupting it’s interaction with PMCA-2b
◦ …to develop a PDZ binding ligand with improved binding characteristics over an
existing PDZ binding peptide ligand, R2…
◦ …the putative ligand should therefore possess a smaller Kd dissociation constant
binding at nano-molar concentrations – R2 currently binds at micromolar
concentrations.”
Structure and Function
◦ Pizerchio et al proposed a cyclic peptide structure based on the straight chain
C-Termini of PMCA2b but with a cyclic linker – This led to development of R2 ligand
by St. Georges, Royal Holloway, university of Otago, New Zealand and uni. of London
- R2 presented at Association International Conference on Alzheimer's Disease, 2010
◦ Later studies by Li, Spaller et al
have yielded important
thermodynamic data relating
to the effects of alterations in
the ring length and residues on
binding stability
New Ligand J1 (Jamie-1) based on R2 (Ruth-2)
◦ Incorporates
Hexa-Arginine sequence – Cell penetration enhancer
Backbone - based on C-Terminus of PMCA2b
Cyclic ring - with β-Aspartyl linker (previously β-Alanine)
Biotin Marker - to aid in microscopy and assess how much drug penetrates neurones
◦ J1 modeled
In Silico
Left : J1 shown without
Hexa-Arg or Biotin
Above : J1, note shorthand
Hexa-Arginine on left of structure
Molecular docking
◦ A methodology by which a putative ligand, stored as a 3d data model is docked to a
target protein’s active site (also stored as a 3d model).
◦ The software re-arranges the atoms to compute the lowest overall energy by means
of energy minimisation algorithms such as GROMOS or docking scores such as PMF
Argus Lab: PSD-95 protein
with J1 Ligand (yellow)
Scigress: PSD-95 protein
with J1 Ligand (red/blue/grey)
Molecular docking
◦ β-Aspartyl was chosen as a linker as we want to facilitate the bonding between the
β-Asp negative COO- side-chain and a nearby Lysine-98 which has a positive NH3+
side-chain.
◦ This bonding will be ionic providing extra stability to the bound complex
Zeus-PDB: PSD-95 protein
with J1 Ligand (green)
DeepView: PSD-95 protein
with J1 Ligand (red/blue/white)
Lysine 98 (green) – (Zoomed in)
I simulatated the docking of a selection of L- and D- linker ligands in
both alpha and beta- configurations, the results determined what was
synthesised
L- and D- amino acids in alpha and beta config
Some standard amino acids in L-a- config
SPPS, Solid Phase Peptide Synthesis
◦
◦
◦
◦
Original method pioneered by Robert Bruce Merrifield
Essentially synthesis of peptide from C-terminus to the N-terminus, stepwise
First residue is bound to solid beads and is protected
Beads are washed with a solution of the subsequent residue, which is then
washed again to deprotect it. The cycle is repeated with each further residue and
finally washed with TFA (TriFluoracetic Acid to cleave the peptide from the
beads).
◦ Straight chain of J1 can be automated using St. George’s Peptide synthesiser
◦ Linker (ring with β-Asp) will be added through a separate reaction to connect the
side-chains (Lysine-2 and Glutamate-4 to the linker, β-Aspartyl).
Synthesis of J1
◦ Synthesis will be on a Leu Peg-PS resin using temporary α-amino Fmoc
protection and orthogonally protected residues
◦ Residues will be deprotected and reacted with the bridging residue before finally
being deprotected.
◦ The synthesised peptide will be purified using HPLC
◦ The product is then verified using mass spectrometry (MS) and MS2 by Maldi
Orthogonal Protecting group strategy
(Figure 30) - Schematic of the J-1 (Jamie-1) backbone – precursor to cyclic J-1 based on R-2 (Ruth-2).
(From right to left) blue region shows the initial Leucine, Leu-1 bound to the Resin (Fmoc-Leu-PEG-PS)
followed by Orthogonally protected groups (shown in red) on Lys-2(IvDde) and Glu-4(PhiPr) which are
separated by Thr-3. Two conventionally Fmoc coupled residues Leu-5 and Ser-6 precede Biotinylated
Lysine, Lys-7 (shown in Green). Gly-8 and Gly-9 are proceeded by Pbf protected Hexa-Arginine unit (Purple
with Pbf shown in Magenta) which has been acetylated. Two t-Butyl groups are shown in Orange and are
used to protect the hydroxyl of Threonine and Serine.
FMoc deprotection and cleavage
Fmoc protection strategy is stable in acidic conditions and therefore required Basic
condition for cleavage of temporary protection (Piperidine 20% in DMF)
HATU activation of acyl groups during peptide construction
Characterisation of synthesised peptides
HPLC (High performance Liquid Chromatography)
MALDI mass spectrometry
Determine the Kd binding constant
GST (Glutathione-S-Transferase) Pull-down assay used to attain Kd dissociation constant
GST is a marker-tag that can be attached to proteins under study
If binding occurs then the complex will contain marked protein+bound protein
In the presence of J1 we can quantify the potency of binding between J1 and PSD95-PDZ1
Quantifying the effect of J1 on altering excitotoxicity
◦ Utilises a cell line, SHSY-5Y cells derived from 30 year old blastoma stored at
St. Georges, refrigerated at a specific temp. & under CO2 to prevent alkalinisation.
◦ J1 will be added to the SHSY-5Y neuronal cells in presence of excess glutamate
(2mmol) and quantifying the amount of cells that survive.
Immunohistological analysis
◦ Used to study the extent to which J1 internalises within
SHSY-5Y cells and to PSD-95
◦ Makes use of the integrated biotin marker within the structure as a
target for anti-biotin antibody - visualised by microscopy
Biotin+Ab. marked neuronal cells
SHSY-5Y cells cultured in DMEM-F12 buffer
Fluorophores (three Flurophores were used)
◦ DAPI – to label the A-T rich regions of DNA in the Nuclei (for
contrasting and locating cells) FITC-Avidin conjugate to label Biotin
incorporated in PDZ peptides (labels the peptide) TRITC-anti-rabbit
antibody for PSD-95 (labels the PSD-95 target on the membrane)
Jamie-3-Asn
Slide#
Jamie-3-Glu
1A
2A
3A
4A
1B
2B
3B
4B
2.7uM PDZ
peptide
X
X
X
X
X
X
X
X
DAPI (Blue)
X
X
X
X
X
X
X
X
FITCAvidin(Green)
X
X
X
X
X
X
Anti-PSD95
X
X
X
X
X
X
TRITC Antirabbit (Red)
X
X
X
X
X
X
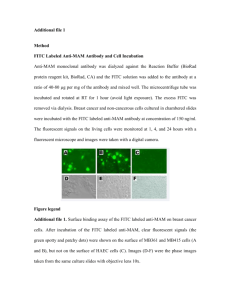
DAPI / FITC / TRITC stained SHSY-5Y cells, Jamie-3 peptide
(x40)
Jamie 3-Glu (DAPI+FITC)
( Slide #1B)
Jamie 3-Glu (DAPI+FITC)
Jamie 3-Glu (DAPI+TRITC)
Negative control slides ( lacking either
FITC {peptide} or TRITC {psd95 target} )
DAPI / FITC / TRITC stained SHSY-5Y cells, Jamie-3-Asn (x63)
Jamie 3-Asn
(DAPI+FITC+TRITC)
Top left = DAPI
Top right =
FITC (peptide)
Bottom left = TRITC
Bottom right =
Composite image
(DAPI+FITC+TRITC)
showing good
co-localisation
(orange and yellow) of
Jamie-3-Asn (green)
and target PSD-95
(red)
FITC / TRITC stained SHSY-5Y cells, Jamie-3 Asn (x63)
Composite image of Jamie 3-Asn
FITC+TRITC but excluding DAPI.
It shows co-localisation (orange
and yellow) of the Jamie-3-Asn
peptide with its PSD-95 target.
Jamie-3-Asn was stained with FITC
and the PSD-95 target with
Anti-PSD95 TRITC the co-localisation
is the combination of the two colours
Slide #3A
FITC / TRITC stained SHSY-5Y cells, Jamie-3 Asn (x63)
Composite image of Jamie 3Asn FITC+TRITC+DAPI
It shows co-localisation (orange
and yellow) of the Jamie-3-Asn
peptide with its PSD-95 target.
Jamie-3-Asn was stained with
FITC and the PSD-95 target with
Anti-PSD95 TRITC the colocalisation is the combination of
the two colours
Slide #3A
PDZ domains
◦ PDZ domains are important protein-protein cell signaling mediators
◦ Implicated in a variety of cellular processes not just neurological
◦ Important drug target for novel drugs
Molecular modeling and In Silico methods
◦ Molecular modeling and docking is extremely useful method of simulating
thermodynamic stability and binding between ligands and proteins
◦ Data can be used to select a putative ligand prior synthesis vs the traditional
combinatorial approach
Immunohistochemical analysis
◦
◦
◦
Jamie-3-Asn pdz binding peptide showed good co-localisation of peptide with PSD-95
Jamie-3-Glu showed some co-localisation but was less consistent across repeated imaging
Results consistent with MTT assay, in which Jamie-3-Asn pdz binding peptide performed
better at mitigating excitotoxic damage than Jamie-3-Glu
Implications
◦ More favorable binding characteristics than R2 - further development may yield a
useful pharmacological agent that could be administered to slow down neurological
degeneration in diseases such as Alzeimers, Parkinsons, epilepsy and stroke.
References used
1. Harris BZ, Lim WA. Mechanism and role of PDZ domains in signaling complex
assembly. Journal of Cell Science, 2010; 114: 3219-3231
2. Cui H, Hayashi A, Sun HS, Belmares MP, Cobey C, Phan T et al. PDZ Protein
Interactions Underlying NMDA Receptor-Mediated Excitotoxicity and
Neuroprotection by PSD-95 Inhibitors. The Journal of Neuroscience, 2007; 27(37):
9901-9915
3. Li T, Saro D, Spaller MR. Thermodynamic profiling of conformationally constrained
cyclic ligands for the PDZ domain. Bioorganic & Medicinal Chemistry Letters, 2004;
14(6): 1385-1388
4. Austen BM, Duberley K, Turner P, Empson R. Cyclic hexa-arg PDZ-binding peptides
that bind PSD95 inhibit glutamate-mediated toxicity. Alzheimer's Association
International Conference on Alzheimer's Disease 2010, 6(4): Supp. 562
Software used (in order of images shown)
1.
2.
3.
4.
Argus Lab by Mark Thompson
Scigress explorer by Fujitsu Siemens
Zeus PDB viewer by Jamie al-nasir
DeepView/Swiss-PDB viewer