Theme 1
advertisement

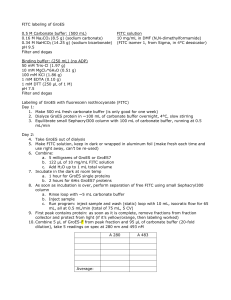
Theme 1 Project 1: Supervisor: Molecular Design Redesigning Protein Interactions through Chemical Modification of the Protein Surface Dr. Peter B. Crowley, School of Chemistry Project Summary: Protein-protein interactions are central to organisation and function in the living cell. We are interested in understanding the determinants of protein recognition and binding affinity. The aim of this project is to investigate how small molecule modifications of the protein surface can influence that protein’s interaction behaviour. Proteins labelled with fluorescent dyes are used extensively in protein interaction studies both in vivo and in vitro. Frequently, such dyes (e.g. FITC) are bulky, predominantly hydrophobic molecules. Considering that the labelling procedure is often non-specific (via amino groups in the case of FITC) we Figure 1. FITC binding site in hypothesize that the surface of the labelled protein may be 1flr. Aromatic side chains are significantly altered, resulting in non-native protein depicted in sticks. FITC is green. interactions. Indeed several crystal structures of protein-FITC complexes (PDB entries: 1n0s, 1t66, 1flr) reveal that FITC is accommodated in hydrophobic binding pockets (see Figure 1).1 Therefore, we propose to rigorously test the effect of labelling on protein interaction behaviour. To make the problem more readily tractable we propose to study a dimer system. The test molecule is Azurin, a structurally well-characterised -sheet protein, which can Figure 2. FITC-Azurin uptake in invade mammalian cells.2 The crystal structure of Azurin breast cancer cell line MCF-7. reveals a dimeric arrangement assembled via the conserved Nuclei are stained with DAPI. surface hydrophobic patch.3 The architecture of the dimer interface makes Azurin an interesting test molecule for these studies. Using selective labelling via surface exposed cysteine residues,4 bulky hydrophobic molecules (FITC analogues) will be introduced at different sites in and around the hydrophobic patch. Biophysical methods including size exclusion chromatography, NMR spectroscopy and X-ray crystallography will be used to assess the effect on dimer formation. This project is integral to our investigation of protein interactions. We aim to make progress in the area of protein-ligand interactions.5 It is envisaged that the successful completion of the project will lead to at least one publication as well as providing preliminary data for future grant applications (e.g. RFP). In addition, the results of the in vitro experiments will be used to inform/interpret our cell uptake studies (Figure 2). References: [1] Terzyan et al. J. Mol. Biol. 2004, 339, 1141-1151. [2] Yaamada et al. Proc. Natl. Acad. Sci. USA 2004, 101, 4770-4775. [3] van Amsterdam et al. Nat. Struct. Biol. 2002, 9, 48-52. [4] Simon et al. Cell 2007, 128, 1003–1012. [5] Crowley et al. Chem. Bio. Chem. 2008, 9, 1029-1033.




![Anti-Integrin alpha 2 antibody [AK7] (FITC) ab30486](http://s2.studylib.net/store/data/012090999_1-09f9400fb7ae466a855733f47c2e099e-300x300.png)