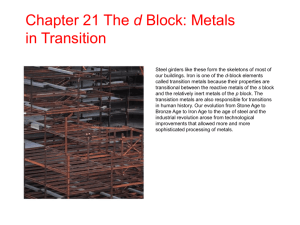
Computational modeling of the copper
advertisement

Computational modeling of the copper-binding domain of Alzheimer’s Amyloid-β peptide – What is the fourth oxygen-donor ligand? Yogita Mantri, Marco Fioroni and Mu-Hyun Baik* Department of Chemistry, Indiana University, Bloomington, IN, 47405, USA The 42-residue Amyloid-β peptide of Alzheimer’s disease is widely believed to bind copper, which subsequently catalyzes the reduction of dioxygen to hydrogen peroxide. Only the first 16 amino acids appear to be involved in copper-binding, while EPR studies suggest that the coordination environment around copper is of the type 3N1O. The nitrogen-donor ligands are most likely His6, His13 and His14. The identity of the fourth, oxygen-donor ligand is controversial at present. Knowledge of all four ligands is crucial for understanding the catalytic mechanism of this complex. We aim to identify the mystery ligand by comparative modeling of all possible O-donor ligands among the first 16 residues of Aβ42 in terms of copper-binding energies, and propose the fourth ligand to copper to be Glu11. The computational methodology involved high-level DFT calculations on simplified models of the copper-binding site and classical molecular dynamics simulations of the full-length peptides. While absolute binding energies are difficult to predict, the relative energies of the conformers are likely more reliable. Not surprisingly, the quantum mechanical models predict the anionic ligands Asp and Glu to have the strongest binding energies. Addition of the peptide distortion energies allows for further refinement and slightly shuffles the relative rankings, with the most favorable binding energy predicted for Glu11, followed closely by Tyr10, which has been previously proposed as the Odonor ligand. Figure 1. Representative conformation of the Aβ42-CuII complex with Glu11 as the O-donor ligand. The ligands to copper (His6, His13, His14 and Glu11) are shown in stick form.