jam12523-sup-0001-TableS1-2
advertisement

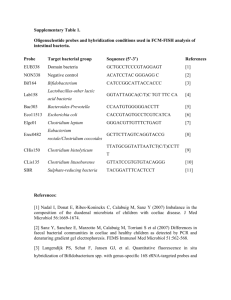
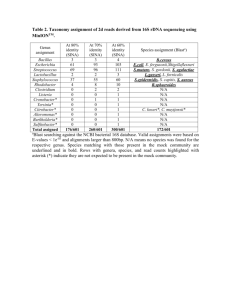
Microbial community structure of wastewater treatment subjected to high mortality rate due to ozonation of return activated sludge Siavash Isazadeh, Pinar Ozdural Ozcer, and Dominic Frigon* Department of Civil Engineering and Applied Mechanics, McGill University, 817 Sherbrooke Street West, Montreal, QC, H3A 0C3, Canada * Corresponding author Email: dominic.frigon@mcgill.ca Tel: +1-514-398, 2475 FAX: +1-514-398, 7361 SUPPLEMENTARY MATERIAL Number of tables=2 1 Table S1. Physical and operational characteristic of two pilot-scale reactor. Parameters Control RAS-Ozonated Influent Flow (m3/day) 1.81±0.09 1.89±0.11 Volume of aeration tank (m3) 1.045 1.015 Volume of clarifier (m3) 0.705 0.697 Aerated SRT (d) 6.06 7.17 RAS Flow/Influent Flow 1.5 1.3 RAS suspended solids (mg/L) 3,040 3,040 Ozone contact time (min) - 45 Ozone flow rate (L/h) - 2-6 2 Table S2. PCR primers and FISH probes used in this study. Probes and primers Label Sequence (5’–3’) Binding position b Target Group FA %c Annealing temperature References FISH EUB338a FITC GCTGCCTCCCGTAG 16S (338–355) Most Bacteria 0 – 50 NAd (Amann 1990) a GAGT Other Bacteria not detected by EUB338-II FITC GCAGCCACCCGTA 16S (338–355) 0 – 50 NA (Daims 1999) GGTGT EUB338 Other Bacteria not detected by EUB338-IIIa FITC GCTGCCACCCGTAG 16S (338–355) 0 – 50 NA (Daims 1999) GTGT EUB338 ALF968 Cy3 GGTAAGGTTCTGCG 16S (968 - 985) Alphaproteobacteria 35 NA (Neef 1997) CGTT BET42a Cy5 GCCTTCCCACTTCG 23S (1027–1043) Betaroteobacteria 35 NA (Manz 1992) TTT GCCTTCCCACATCG Competitor for BET42a 35 NA TTT GAM42a Cy3 GCCTTCCCACATCG 23S (1027–1043) Gamaproteobacteria 35 NA (Manz 1992) TTT GCCTTCCCACTTCG Competitor for GAM42a 35 NA AGTTAGCCGGTGCT 16S (495–512) (Lucker et al. DELTA495a Cy5 TTT Deltaproteobacteria 35 NA TCCT 2007) AGTTAGCCGGTGCT Competitor for DELTA495a 35 NA TCTT CFB563 Cy5 GGACCCTTTAAACC 16S (563–580) Flavobacteria 20 NA CAAT LGC354a Cy5 TGGAAGATTCCCTA 16S (354–371) Firmicutes NA (Meier et al. 1999) CTGC HGC69a Cy3 TATAGTTACCACCG 23S (1901–1918) Actinobacteria 20 NA (Roller 1994) CCGT Pyrosequencing CCTACGGGAGGCA 16S (341-357) (Muyzer et al. 341F NA Most Bacteria NA NA GCAG CCGTCAATTCCTTT 16S (891-907) 907R NA Most Bacteria NA NA (Lane 1993) et al. 1985) GAGTTT Cloning ACTCCTACGGGAG 16S (338-354) Non-EUB338F NA Most Bacteria NA 56 (Amann 1990) GCAGCAG TACGTGTGAAGCCC Meth1215Re NA 16S (-1199-1215) Methylophilaceae NA 56 this study TGGC AATACGACTCACTA T7 Universal Primer NA TAG GTTTTCCCAGTCAC M13 Universal primer NA 16s (20-41) GAC a EUB338, EUB338-II, EUB338-III were used in the mixture called EUB mix b rRNA target site (Escherichia coli numbering) c FA, formamide concentration in the hybridization buffer. For salt concentration in wash buffer see(Nielsen and Daims 2009) d NA: Not Applicable e Meth1215R coverage (91% of Methylophilaceae) and specificity(88%Methylophilaceae) based on ribosomal database project ( RDP Release 10, accessed in June 2012) 3 References Amann, R.I., 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Applied and Environmental Microbiology 56(6), 1919. Daims, H., 1999. The domain-specific probe EUB338 is insufficient for the detection of all bacteria: Development and evaluation of a more comprehensive probe set. Systematic and Applied Microbiology 22(3), 434. Lane, D.J., Pace, B., Olsen, G.J., Stahl, D.A., Sogin, M.L., Pace, N.R., 1985. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proceedings of the National Academy of Sciences 82(20), 6955-6959. Lucker, S., Steger, D., Kjeldsen, K.U., MacGregor, B.J., Wagner, M., Loy, A., 2007. Improved 16S rRNA-targeted probe set for analysis of sulfate-reducing bacteria by fluorescence in situ hybridization. Journal of Microbiological Methods 69(3), 523-528. Manz, W., 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Systematic and Applied Microbiology 15(4), 593. Meier, H., Amann, R., Ludwig, W., Schleifer, K.H., 1999. Specific Oligonucleotide Probes for in situ Detection of a Major Group of Gram-positive Bacteria with low DNA G+C Content. Systematic and Applied Microbiology 22(2), 186-196. Muyzer, G., de Waal, E.C., Uitterlinden, A.G., 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reactionamplified genes coding for 16S rRNA. Applied and Environmental Microbiology 59(3), 695-700. Neef, A., 1997. Anwendung der in situ Einzelzell-Identifizierung von Bakterien zur Populationsanalyse in komplexen mikrobiellen Biozönosen, Technische Universität München Munich, Germany. Nielsen, P.H., Daims, H., 2009. FISH handbook for biological wastewater treatment: identification and quantification of microorganisms in activated sludge and biofilms by FISH, IWA Publishing, London, UK. Roller, C., 1994. In situ probing of Gram-positive bacteria with high DNA G+ C content using 23S rRNA-targeted oligonucleotides. Microbiology 140(10), 2849. 4