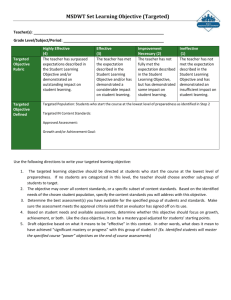
Slides
advertisement

Targeted Maximum Likelihood
Super Learning
Application to assess effects in
RCT,Observational Studies, and
Genomics
Mark van der Laan
works.bepress.com/mark_van_der_laan
Division of Biostatistics,
University of California, Berkeley
Workshop Brad Efron, December 2009
Outline
•
•
•
•
•
•
Super Learning and Targeted Maximum
Likelihood Learning
Causal effect in observational studies
Causal effect in RCTs
Variable importance analysis in
Genomics
Multiple testing
Case Control data
Motivation
• Avoid reliance on human art and parametric
models
• Adapt the model fit to the data
• Target the fit to the parameter of interest
• Statistical Inference
TMLE/SL
Targeted Maximum Likelihood coupled with Super Learner methodology
TMLE/SL Toolbox
Targeted effects
•
Effect of static or dynamic multiple time point treatments
(e.g. on survival time)
•
Direct and Indirect Effects
•
Variable importance analysis in genomics
Types of data
•
Point treatment
•
Longitudinal/Repeated Measures
•
Censoring/Missingness/Time-dependent confounding.
•
Case-Control
•
Randomized clinical trials and observational data
4
Two-stage Methodology: SL/TMLE
1. Super Learning
• Works on a library of model fits
• Builds data-adaptive composite
model by assigning weights
• Weights are optimized based on lossfunction specific cross-validation to
guarantee best overall fit
2. Targeted Maximum
Likelihood Estimation
• Zooms in on one aspect of the
model fit—the target
• Removes bias for the target.
Loss-Based Super Learning in
Semiparametric Models
• Allows one to combine many data adaptive (e.g.)
MLEs into one improved MLE.
• Grounded by oracle results for loss-function
based cross-validation (vdL&D, 2003). Loss
function needs to be bounded.
• Performs asymptotically as well as best (oracle)
weighted combination, or achieves parametric
rate of convergence.
Super Learner Flow Chart in
Prediction
Super Learner Prediction
Targeted Maximum Likelihood
Estimation Flow Chart
Inputs
The model is a set of possible
probability distributions
of the data
Initial P-estimator of the
probability distribution
of the data: Pˆ
Model
Pˆ
User Dataset
ˆ
P*
Targeted P-estimator of the
probability distribution
of the data
O(1), O(2),
… O(n)
PTRUE
Observations
True probability distribution
Target feature map: Ψ( )
Ψ(PTRUE)
ˆ
Ψ(P)
Initial feature estimator
ˆ
Ψ(P*)
Targeted feature estimator
Target feature values
True value
of the target feature
Target Feature
better estimates are closer to ψ(PTRUE)
Targeted Maximum Likelihood
• MLE/SL aims to do good job of estimating whole
density
• Targeted MLE aims to do good job at parameter of
interest
• General decrease in bias for parameter of
Interest
• Fewer false positives
• Honest p-values, inference, multiple testing
(Iterative) Targeted MLE
1.
^
Identify optimal strategy for “stretching” initial P
–
2.
Small “stretch” -> maximum change in target
Given strategy, identify optimum amount of stretch by
MLE
^
3.
Apply optimal stretch to P using optimal stretching
function -> 1st-step targeted maximum likelihood
estimator
4.
Repeat until the incremental “stretch” is zero
–
5.
Some important cases: 1 step to convergence
Final probability distribution solves efficient influence
curve equation
(Iterative) T-MLE is double robust & locally efficient
Example: Targeted MLE of
Causal effect of point
treatment on outcome
Impact of Treatment on Disease
Likelihood of Point Treatment
with Single Endpoint Outcome
•
•
•
•
•
Draw baseline characteristics
Draw treatment
Draw missing indicator
If not missing, draw outcome
Counterfactual outcome distributions
defined by intervening on treatment and
enforcing no missingness
• Causal effects defined as user supplied
function of these counterfactual
TMLE for Average Causal Effect
• Observe predictors W, treatment A, missingness indicator
Delta, and outcome Y:
• Target is additive causal effect: EY(1)-Y(0)
• Regress Y on treatment A and W and Delta=1 (e.g. Super
Learning), and add clever covariate
where
– Then average regression over W for fixed treatment a: EnYa
• Evaluate average effect: EnY1-EnY0
TMLE is Collaborative Double
Robust
• Suppose the initial fit minus true outcome regression is
only a function of W through S
• Suppose the treatment mechanism adjusts correctly for a
set of variables that includes S
• Then, the Targeted MLE is consistent.
• Thus the treatment mechanism only needs to adjust for
covariates whose
effect has not been captured by the
initial fit yet.
• Formally,
TMLE/SL: more accurate information from less data
Simulated Safety Analysis of Epogen (Amgen)
Example: Targeted MLE in
RCT
Impact of Treatment on Disease
The Gain in Relative Efficiency in RCT is function
of Gain in R^2 relative to unadjusted estimator
• We observe (W,A,Y) on each unit
• A is randomized, P(A=1)=0.5
• Suppose the target parameter is additive causal
effect EY(1)-Y(0)
• The relative efficiency of the unadjusted
estimator and a targeted MLE equals 1 minus
the R-square of the regression
0.5 Q(1,W)+0.5 Q(0,W), where Q(A,W) is the
regression of Y on A,W obtained with targeted
MLE.
TMLE in Actual Phase IV RCT
• Study: RCT aims to evaluate safety based on
mortality due to drug-to-drug interaction among
patients with severe disease
• Data obtained with random sampling from
original real RCT FDA dataset
• Goal: Estimate risk difference (RD) in survival at
28 days (0/1 outcome) between treated and
placebo groups
TMLE in Phase IV RCT
Estimate
p-value (RE)
Unadjusted
TMLE
0.034
0.043
0.085 (1.000)
0.009 (1.202)
• TMLE adjusts for small amount of empirical confounding
(imbalance in AGE covariate)
• TMLE exploits the covariate information to gain in
efficiency and thus power over unadjusted
• TMLE Results significant at 0.05
TMLE in RCT: Summary
• TMLE approach handles censoring and improves
efficiency over standard approaches
– Measure strong predictors of outcome
• Implications
–
–
–
–
–
Unbiased estimates with informative censoring
Improved power for clinical trials
Smaller sample sizes needed
Possible to employ earlier stopping rules
Less need for homogeneity in sample
• More representative sampling
• Expanded opportunities for subgroup analyses
Targeted MLE
Analysis of Genomic Data
Biomarker discovery, Impact of
mutations on disease, or response to
treatment
The Need for Experimentation
• Estimation of Variable Importance/Causal
Effect requires assumption not needed for
prediction
• “Experimental Treatment Assignment”
(ETA)
– Must be some variation in treatment variable A
within every stratum of confounders W
• W must not perfectly predict/determine A
• g(a|W)>0 for all (a,W)
Biomarker Discovery: HIV
Resistance Mutations
• Goal: Rank a set of genetic mutations based on
their importance for determining an outcome
– Mutations (A) in the HIV protease enzyme
• Measured by sequencing
– Outcome (Y) = change in viral load 12 weeks after
starting new regimen containing saquinavir
• How important is each mutation for viral
resistance to this specific protease inhibitor
drug?
– Inform genotypic scoring systems
Stanford Drug Resistance
Database
• All Treatment Change Episodes (TCEs) in the
Stanford Drug Resistance Database
– Patients drawn from 16 clinics in Northern CA
Baseline
Viral Load <24 weeks
12 weeks
TCE (Change >= 1 Drug)
Final
Viral Load
Change in Regimen
• 333 patients on saquinavir regimen
Parameter of Interest
• Need to control for a range of other covariates W
– Include: past treatment history, baseline clinical
characteristics, non-protease mutations, other drugs
in regimen
• Parameter of Interest Variable Importance
ψ = E[E(Y|Aj=1,W)-E(Y|Aj=0,W)]
– For each protease mutation (indexed by j)
Parameter of Interest
• Assuming no unmeasured confounders
(W sufficient to control for confounding)
Causal Effect is same as W-adjusted
Variable Importance
E(Y1)-E(Y0)=E[E(Y|A=1,W)-E(Y|A=0,W)]= ψ
– Same advantages to T-MLE
Targeted Maximum Likelihood Estimation of the Adjusted Effect of HIV
Mutation on Resistance to Lopinavir
mutation
p50V
p82AFST
p54VA
p54LMST
p84AV
p46ILV
p48VM
p47V
p32I
p90M
p82MLC
p84C
p33F
p53LY
p73CSTA
p24IF
p10FIRVY
p71TVI
p30N
p88S
p88DTG
p36ILVTA
p20IMRTVL
p23I
p16E
p63P
score
20
20
11
11
11
11
10
10
10
10
10
10
5
3
2
2
2
2
0
0
0
0
0
0
0
0
estimate
1.703
0.389
0.505
0.369
0.099
0.046
0.306
0.805
0.544
0.209
1.610
0.602
0.300
0.214
0.635
0.229
-0.266
0.019
-0.440
-0.474
-0.426
0.272
0.178
0.822
0.239
-0.131
95% CI
( 0.760, 2.645)*
( 0.084, 0.695)*
( 0.241, 0.770)*
( 0.002, 0.735)*
(-0.130, 0.329)
(-0.222, 0.315)
(-0.162, 0.774)
( 0.282, 1.328)*
( 0.312, 0.777)*
(-0.058, 0.476)
( 1.330, 1.890)*
( 0.471, 0.734)*
(-0.070, 0.669)
(-0.266, 0.695)
( 0.278, 0.992)*
(-0.215, 0.674)
(-0.522, -0.011)*
(-0.243, 0.281)
(-0.853, -0.028)*
(-0.840, -0.108)*
(-0.842, -0.010)*
(-0.001, 0.544)
(-0.111, 0.467)
(-0.050, 1.694)
(-0.156, 0.633)
(-0.392, 0.131)
Stanford mutation score, http://hivdb.stanford.edu, accessed September, 1997
Multiple Testing:
Combining Targeted MLE
with Type-I Error Control
Hypothesis Testing Ingredients
Data (X1,…,Xn)
• Hypotheses
• Test Statistics
• Type I Error
• Null Distribution
• Marginal (p-values) or
• Joint distribution of the test statistics
• Rejection Region
• Adjusted p-values
Type I Error Rates
• FWER: Control the probability of at least one
Type I error (Vn): P(Vn > 0) ·
• gFWER: Control the probability of at least k
Type I errors (Vn): P(Vn > k) ·
• TPPFP: Control the proportion of Type I errors
(Vn) to total rejections (Rn) at a user defined
level q: P(Vn/Rn > q) ·
• FDR: Control the expectation of the proportion of
Type I errors to total rejections: E(Vn/Rn) ·
Multivariate Normal Null
Distribution
• Suppose null hypotheses involve testing of
target parameters H_0: psi(j)<=0
• We estimate target parameters with TMLE, and use t-statistic for testing
• T-MLE as vector is asymptotically linear
with known influence curve IC
• Valid joint null distribution for multiple
testing is N(0,Sigma=E IC^2)
• Null distr can be inputted in any MTP
(Dudoit, vdL, 2009, Springer)
GENERAL JOINT NULL
DISTRIBUTION
Let Q0j be a marginal null distribution so that
for j in set S0 of true nulls
Q0j-1Qnj(x)> x, for all x
where Qnj is the j-th marginal distribution of
the true distribution Qn(P) of the test
statistic vector Tn.
JOINT NULL DISTRUTION
We propose as null distribution the
distribution Q0n of
Tn*(j)=Q0j-1Qnj(Tn(j)), j=1,…,J
This joint null distribution Q0n(P) does indeed
satisfy the wished multivariate asymptotic
domination condition in (Dudoit, van der
Laan, Pollard, 2004).
BOOTSTRAP BASED JOINT NULL
DISTRIBUTION
We estimate this null distribution Q0n(P) with
the bootstrap analogue:
Tn#(j)=Q0j-1Qnj#(Tn#(j))
where # denotes the analogue based on
bootstrap sample O1#,..,On# of an
approximation Pn of the true distribution P.
Case-Control Weighted Targeted MLE
• Case-control weighting in targeted MLE successfully
maps an estimation method designed for prospective
sampling into a method for case-control sampling.
• This technique relies on knowledge of the true
prevalence probability P(Y=1)=q0 to eliminate the
bias of the case-control sampling design.
• The procedure is double robust and locally efficient.
It produces efficient estimators when its prospective
sample counterpart is efficient.
Comparison to Existing Methodology
Case-control weighted targeted MLE differs from other approaches as it
can estimate any type of parameter, incorporates q0, and is double robust
and locally efficient.
Case-Control Weighted Targeted MLE
Simulation Results
• We showed striking
improvements in efficiency and
bias in our case-control weighted
method versus the IPTW
estimator (Mansson 2007,
Table results for a sample of 500 cases and 1000 controls
taken from a population of 120,000 where q0 = 0.035
Robins 1999), which does not
utilize q0.
• Our complete simulation results
bolster our theoretical arguments
that gains in efficiency and
reductions in bias can be
obtained by having known q0 and
using a targeted estimator.
Closing Remarks
• True knowledge is embodied by semi or nonparametric models
• Semi-parametric models require fully automated
state of the art machine learning (super learning)
• Targeted bias removal is essential and is
achieved by targeted MLE
• Statistical inference is now sensible
• The machine learning algorithms are (super)
efficient for the target parameters.
Closing Remarks
• (RC) Clinical Trials and Observational
Studies can be analyzed with TMLE.
• TMLE outperforms current standards in
analysis of clinical trials and observational
studies, including double robust methods
• It is the only targeted method that is
collaborative double robust, efficient, and
naturally incorporates machine learning
Acknowledgements
• UC Berkeley
–
–
–
–
–
–
–
–
–
Oliver Bembom
Susan Gruber
Kelly Moore
Maya Petersen
Dan Rubin
Cathy Tuglus
Sherri Rose
Michael Rosenblum
Eric Polley
– P.I. Ira Tager (Epi).
• Stanford Univ.
– Robert Shafer
• Kaiser: Dr. Jeffrey
Fessels.…
• FDA: Thamban
Valappil, Greg Soon,
• Harvard:
David Bangsberg,
Victor DeGruttolas
References
•
•
•
•
•
•
Oliver Bembom, Maya L. Petersen , Soo-Yon Rhee , W. Jeffrey Fessel , Sandra E.
Sinisi, Robert W. Shafer, and Mark J. van der Laan, "Biomarker Discovery Using
Targeted Maximum Likelihood Estimation: Application to the Treatment of
Antiretroviral Resistant HIV Infection" (August 2007). U.C. Berkeley Division of
Biostatistics Working Paper Series. Working Paper 221.
http://www.bepress.com/ucbbiostat/paper221
Mark J. van der Laan and Susan Gruber, "Collaborative Double Robust Targeted
Penalized Maximum Likelihood Estimation" (April 2009). U.C. Berkeley Division of
Biostatistics Working Paper Series. Working Paper 246,
http://www.bepress.com/ucbbiostat/paper246
Mark J. van der Laan, Eric C. Polley, and Alan E. Hubbard, "Super Learner" (July
2007). U.C. Berkeley Division of Biostatistics Working Paper Series. Working Paper
222.
http://www.bepress.com/ucbbiostat/paper222
Mark J. van der Laan and Daniel Rubin, "Targeted Maximum Likelihood Learning"
(October 2006). U.C. Berkeley Division of Biostatistics Working Paper Series.
Working Paper 213.
http://www.bepress.com/ucbbiostat/paper213
Oliver Bembom, Mark van der Laan (2008), A practical illustration of the importance
of realistic individualized treatment rules in causal inference, Electronic Journal of
Statistics.
Mark J. van der Laan, "Statistical Inference for Variable Importance" (August 2005).
U.C. Berkeley Division of Biostatistics Working Paper Series. Working Paper 188.
http://www.bepress.com/ucbbiostat/paper188
References
•
•
•
•
•
•
Oliver Bembom, Maya L. Petersen , Soo-Yon Rhee , W. Jeffrey Fessel , Sandra E.
Sinisi, Robert W. Shafer, and Mark J. van der Laan, "Biomarker Discovery Using
Targeted Maximum Likelihood Estimation: Application to the Treatment of
Antiretroviral Resistant HIV Infection" (August 2007). U.C. Berkeley Division of
Biostatistics Working Paper Series. Working Paper 221.
http://www.bepress.com/ucbbiostat/paper221
Mark J. van der Laan, Eric C. Polley, and Alan E. Hubbard, "Super Learner" (July
2007). U.C. Berkeley Division of Biostatistics Working Paper Series. Working Paper
222.
http://www.bepress.com/ucbbiostat/paper222
Mark J. van der Laan and Daniel Rubin, "Targeted Maximum Likelihood Learning"
(October 2006). U.C. Berkeley Division of Biostatistics Working Paper Series.
Working Paper 213.
http://www.bepress.com/ucbbiostat/paper213
Yue Wang, Maya L. Petersen, David Bangsberg, and Mark J. van der Laan,
"Diagnosing Bias in the Inverse Probability of Treatment Weighted Estimator
Resulting from Violation of Experimental Treatment Assignment" (September 2006).
U.C. Berkeley Division of Biostatistics Working Paper Series. Working Paper 211.
http://www.bepress.com/ucbbiostat/paper211
Oliver Bembom, Mark van der Laan (2008), A practical illustration of the importance
of realistic individualized treatment rules in causal inference, Electronic Journal of
Statistics.
Mark J. van der Laan, "Statistical Inference for Variable Importance" (August 2005).
U.C. Berkeley Division of Biostatistics Working Paper Series. Working Paper 188.
http://www.bepress.com/ucbbiostat/paper188
Collaborative T-MLE: Building the Propensity
Score Based on Outcome Data
• Initial outcome regression based on super learning
• Construct rich set of one dimensional dimension
reductions of W, that will be used as main terms below
• Select main terms in propensity score using forward
selection based on emp. fit (e.g, loglik) of T-MLE
• If no main term increases emp. fit of TMLE, then carry
out T-MLE update to update initial outcome regression
• Proceed to generate a sequence of T-MLE’s using
increasingly nonparametric treatment mechanisms
• Select the wished T-MLE with cross-validation
The Likelihood for Right
Censored Survival Data
• It starts with the marginal probability distribution of the
baseline covariates.
• Then follows the treatment mechanism.
• Then it follows with a product over time points t
• At each time point t, one writes down likelihood of
censoring at time t, death at time t, and it stops at first
event
• Counterfactual survival distributions are obtained by
intervening on treatment, and censoring.
• This then defines the causal effects of interest as
parameter of likelihood.
TMLE with Survival Outcome
• Suppose one observes baseline covariates, treatment,
and one observes subject up till end of follow up or
death:
• One wishes to estimate causal effect of treatment A on
survival T
• Targeted MLE uses covariate information to adjust for
confounding, informative drop out and to gain efficiency
TMLE with Survival Outcome
• Target ψ1(t0)=Pr(T1>t0) and ψ0(t0)=Pr(T0>t0) – thereby
target treatment effect, e.g.,
1) Difference: Pr(T1>t0) - Pr(T0>t0), 2) Log RH:
• Obtain initial conditional hazard fit (e.g. super learner for
discrete survival) and add two time-dependent
covariates
– Iterate until convergence, then use updated conditional hazard
from final step, and average corresponding conditional survival
over W for fixed treatments 0 and 1
TMLE analogue to log rank test
• The parameter,
corresponds with Cox ph parameter, and
thus log rank parameter
• Targeted MLE targeting this parameter is
double robust
TMLE in RCT with Survival Outcome
Difference at Fixed End Point
Independent Censoring
% Bias
Power
95%
Coverage
Relative
Efficiency
<1%
0.79
0.95
1.00
TMLE
<1%
0.91
TMLE: gain in power over KM
0.95
1.44
KM
Informative Censoring
% Bias
Power
95%
Coverage
Relative
Efficiency
13%
0.88
0.92
1.00
TMLE
<1%
TMLE: unbiased
0.92
0.95
1.50
KM
TMLE in RCT with survival outcome:
Log rank analogue
Independent Censoring
%
Bias
Power
95%
Coverage
Relative
Efficiency
Log rank
<2%
0.13
0.95
1.00
TMLE (correct λ)
<1%
0.22
0.95
1.48
TMLE (mis-spec λ)
<1%
0.19
0.95
1.24
TMLE: gain in power over log rank
Informative Censoring
% Bias
Power
Log rank
32%
0.20*
0.93
1.00
TMLE (correct λ, correct G)
<1%
0.18
0.95
1.44
TMLE (mis-spec λ, correct G)
<1%
0.15
0.95
1.24
TMLE: unbiased
95%
Relative
Coverage Efficiency
Tshepo Study
The Tshepo Study is an open-label,
randomized, 3x2x2 factorial design
study conducted at Princess Marina
Hospital in Gaborone, Botswana to
evaluate the efficacy, tolerability, and
development of drug resistance of six
different first-line cART regimens
Analysis Team: Mark van der Laan, Kelly Moore, Ori Stitelman,
Victor De Gruttola
Tshepo Study: C. William Wester, Ann Muir Thomas, Hermann
Bussmann, Sikhulile Moyo, Joseph Makhema, Tendani
Gaolathe, Vladimir Novitsky2, Max Essex,
Victor deGruttola, and Richard G. Marlink
Tshepo Study
• Causal Effect of treatment NVP/EFV on time till
death, time till virologic failure/dealth/treatment
discontinuation, among other endpoints.
• Effect modification by baseline CD4 and Gender
• Causal effect modification by baseline CD4
Simulation Study
• Generated 500 data sets of 500 observations
each per scenario.
• Varied How Well Hazard Was Specified
(Well/Badly)
• Varied Level of ETA Violation
(Low/Med/High)
• Observed performance of: IPCW, G-comp,
Double Robust Augm IPCW, TMLE, C-TMLE
and TMLE excluding the variable that causes
the eta violation in the treatment mechanism.
MSE And Relative Efficiency of
Estimating P(TA=1>t0)
Low ETA
Medum ETA
High ETA
Method
Well
Specified λ
Badly
Specified λ
Well
Specified λ
Badly
Specified λ
Well
Specified
λ
Badly
Specified λ
IPCW
0.00086
(1.1)
0.00086
(1.1)
0.00117
(1.2)
0.00117
(1.2)
0.00698
(1.9)
0.00698
(1.9)
0.00071
(0.9)
0.00085
(1.1)
0.00086
(1.1)
0.00088
(1.1)
0.00073
(0.7)
0.00106
(1.1)
0.00161
(1.6)
0.00116
(1.2)
0.00071
(0.2)
0.00455
(1.3)
0.00231
(0.6)
0.00819
(2.3)
TMLE
0.00085
(1.1)
0.00088
(1.1)
0.00105
(1.1)
0.00111
(1.1)
0.00347
(1.0)
0.00375
(1.0)
TMLE
w/o ETA
0.00084
(1.0)
0.00088
(1.1)
0.00083
(0.8)
0.00087
(1.2)
0.00079
(0.2)
0.00085
(0.2)
c-TMLE
0.00085
(1.1)
0.00104
(1.3)
0.00085
(0.9)
0.00122
(1.2)
0.00080
(0.2)
0.00101
(0.3)
G-comp
DR-IPCW
Percent Of Time Influence Curve
Based 95 Percent CI Includes
Truth
Low ETA
Medum ETA
High ETA
Method
Well
Specified λ
Badly
Specified λ
Well
Specified λ
Badly
Specified λ
Well
Specified
λ
Badly
Specified λ
IPCW
98.4%
98.4%
97.2%
97.2%
89.8%
89.8%
Double
Robust
94.2%
95.4%
95.0%
96.4%
94.8%
95.8%
TMLE
94.0%
95.2%
95.4%
95.6%
84.8%
87.0%
TMLE
w/o ETA
94.2%
94.8%
94.4%
95.6%
94.6%
95.0%
c-TMLE
94.6%
93.6%
96.0%
95.0%
94.8%
93.2%
Properties of c-TMLE Algorithm
Low ETA
Method
Mean
Moves
% No
Moves
% ETA
Variable
Medum ETA
High ETA
Well
Specified λ
Badly
Specified λ
Well
Specified λ
Badly
Specified λ
Well
Specified
λ
Badly
Specified λ
2.7
9.1
2.1
7.3
2.9
7.1
68.8%
0.2%
74.2%
6.4%
62.6%
1.6%
27.8%
60.4%
1.2%
6.0%
0.0%
0.4%
Case-Control Weighted Targeted MLE
Data Analysis
Full cohort analysis sample size was 2066.
Case-control analysis was repeated in 100
simulations with 269 cases and 538 randomly
selected controls. “% Rej” indicates average
percent rejected tests among the 100
simulations.
• Nested case-control studies are
quite common, and can be
particularly beneficial in
biomarker studies.
• We simulated a nested casecontrol study within a cohort
study examining the impact of
higher physical activity on death
in an aging population.
• With known q0, we obtained
similar point estimates in the
case-control analysis.
Description of Simulation
– 100 subjects each with one random X (say a SNP’s)
uniform over 0, 1 or 2.
– For each subject, 100 binary Y’s, (Y1,...Y100) generated
from a model such that:
• first 95 are independent of X
• Last 5 are associated with X
• All Y’s correlated using random effects model
– 100 hypotheses of interest where the null is the
independence of X and Yi .
– Test statistic is Pearson’s 2 test where the null
distribution is 2 with 2 df.
Description of Simulation, cont.
– Simulated data 1000 times
– Performed the following MTP’s to control FWER at
5%.
• Bonferroni
• Null centered, re-scaled bootstrap (NCRB) – based on 5000
bootstraps
• Quantile-Function Based Null Distribution (QFBND)
– Results
• NCRB anti-conservative (inaccurate)
• Bonferroni very conservative (actual FWER is 0.005)
• QFBND is both accurate (FWER 0.04) and powerful (10 times
the power of Bonferroni).
Empirical Bayes/Resampling
TPPFP Method
• We also devised another resampling based
multiple testing procedure, controlling the
proportion of false positives to total rejections.
• This procedure involves:
– Randomly sampling a guessed set of true null
hypotheses:
H0(j)~Bernoulli (Pr(H0(j)=1|Tj)=p0f0(Tj)/f(Tj) )
based on the Empirical Bayes model:
Tj|H0=1 ~f0
Tj~f
p0=P(H0(j)=1)
(p0=1
conservative)
– Our joint bootstrap null distribution of test
statistics.
Emp. BayesTPPFP Method
1.
Grab a column T~n from the null distribution
of length M.
2.
Draw a length M binary vector corresponding
to S0n.
3.
For a vector of c values calculate:
4.
Repeat 1. and 2. 10,000 times and average
over iterations.
5.
Choose the c value where P(rn(c) > q)· .
Examples/Simulations
Bacterial Microarray Example
• Airborne bacterial levels in specific cities over a
span of several weeks are being collected and
compared.
• A specific Affymetrics array was constructed to
quantify the actual bacterial levels in these air
samples.
• We will be comparing the average (over 17
weeks) strain-specific intensity in San Antonio
versus Austin, Texas.
420 Airborne Bacterial Levels
17 time points
San Antonio vs Austin
Procedure
Bonferroni
FWE
Augmentation
TPPFP
E.Bayes/Bootstrap
TPPFP
Number Rejected
= 0.05
5
= 0.10
= 0.05
6
= 0.10
14
= 0.05
13
= 0.10
21
11
CGH Arrays and Tumors
in
Mice
• 11 Comparative genomic hybridization (CGH) arrays
from cancer tumors of 11 mice.
•
DNA from test cells is directly compared to
DNA from normal cells using bacterial
artificial chromosomes (BACs), which are
small DNA fragments placed on an array.
•
With CGH:
– differentially labeled test [tumor] and reference [healthy]
DNA are co-hybridized to the array.
– Fluorescence ratios on each spot of the array are
calculated.
– The location of each BAC in the genome is known and
thus the ratios can be compiled into a genome-wide copy
number profile
Plot of Adjusted p-values for 3 procedures vs. Rank of
BAC (ranked by magnitude of T-statistic)
Acknowledgements:
• UC Berkeley
–
–
–
–
–
–
–
–
–
Susan Gruber
Ori Stittelman
Kelly Moore
Maya Petersen
Dan Rubin
Cathy Tuglus
Sherri Rose
Michael Rosenblum
Eric Polley
-P.I. Ira Tager (Epi).
• Stanford: Dr. Robert
Shafer
• Kaiser: Dr. Jeffrey Fessels
• Harvard: Dr. David
Bangsberg
• UCSF: Dr. Steve Deeks
• FDA: Thamban Vallappil,
Greg Soon
TargetDiscovery demo
Targeted Maximum Likelihood in
Semiparametric Regression
• Implementation just involves adding a covariate
h(A,W) to the regression model
h( A,W )
d
d
m( A,W | b ) E m( A,W | b ) W
db
db
– When m(A,W|b) is linear
h( A,W ) AW EA W W
• Requires estimating E(A|W)
– E.g. Expected value of A given confounders W
• Robust: Estimate is consistent if either
– E(A|W) is estimated consistently
– E(Y|A,W) is estimated consistently
Model-based Variable Importance
• When the variable of interest (A) is continuous
- Given Observed Data: O=(A,W,Y)~Po
W*={possible biomarkers, demographics, etc..}
A=W*j (current biomarker of interest)
W=W*-j
- Measure of Importance:
Given :
m( A,W | b ) E p (Y | A a,W ) E p (Y | A 0,W )
Define :
(A) EW *[m( A,W | b )]
(a) EW [m(a,W | b )]
1 n
m(a, Wi | b )
n i 1
If linear :
abE[W ]
Simplest Case (Marginal) :
ab 0
Evaluation of Biomarker methods:
simulation
Minimal List length to obtain all 10 “true” variables
100
List Length
Linear Reg
80
VImp w/LARS
RF1
60
RF2
40
20
0
0
0.1
0.2
0.3
0.4
0.5
0.6
0.7
0.8
0.9
Correlation
• No appreciable difference in ranking by importance measure or p-value
–plot above is with respect to ranked importance measures
• List Length for linear regression and randomForest increase with increasing correlation,
Variable Importance w/LARS stays near minimum (10) through ρ=0.6, with only small
decreases in power
• Linear regression list length is 2X Variable Importance list length at ρ=0.4 and 4X at ρ=0.6
• RandomForest (RF2) list length is consistently short than linear regression but still is 50%
than Variable Importance list length at ρ=0.4, and twice as long at ρ=0.6
• Variable importance coupled with LARS estimates true causal effect and
outperforms both linear regression and randomForest
Example: Biomarker Discovery
HIV resistance mutations
Accounting for ETA violations
The Need for Experimentation
• Estimation of Variable Importance/Causal
Effect requires assumption not needed for
prediction
• “Experimental Treatment Assignment”
(ETA)
– Must be some variation in treatment variable A
within every stratum of confounders W
• W must not perfectly predict/determine A
• g(a|W)>0 for all (a,W)
Stanford Drug Resistance
Database
• All Treatment Change Episodes (TCEs) in
the Stanford Drug Resistance Database
– Patients drawn from 16 clinics in Northern CA
Baseline
Viral Load <24 weeks
12 weeks
TCE (Change >= 1 Drug)
Final
Viral Load
Change in Regimen
• 333 patients on saquinavir regimen
Parameter of Interest
• Need to control for a range of other
covariates W
– Include: past treatment history, baseline
clinical characteristics, non-protease
mutations, other drugs in regimen
• Parameter of Interest: Variable Importance
ψ = E[E(Y|Aj=1,W)-E(Y|Aj=0,W)]
– For each protease mutation (indexed by j)
Unadjusted estimates
Delta-adjusted T-MLE
Realistic Causal Effect of
Physical Activity Level
• Given elderly population (SONOMA),
we wish to establish effect of activity
on 5 year mortality.
• A realistic individualized treatment rule
indexed by activity level a is defined
as, given individuals covariates W,
realistic activity level closest to
assigned level a:
• Causal relative risk:E(Y(d(a))/E(Y(d(0))
Realistic Rules Indexed by 5
Activity Levels
Sparse-Data(ETA)-Bias for
Different Causal Effects
Estimates of Realistic/Intention to
Treat/Static Causal Relative Risks
Evaluation of Biomarker Discovery Methods
Methods
> Univariate Linear Regression
• Importance measure: Coefficient value
with associated p-value
• Measures marginal association
> RandomForest (Breiman 2001)
• Importance measures (no p-values)
RF1: variable’s influence on error rate
RF2: mean improvement in node
splits due to variable
> Variable Importance with TMLE based on
LARS
• Importance measure: causal effect
E p (Y | A a,W ) E p (Y | A 0,W )
• Formal inference, p-values provided
• LARS used to fit initial E[Y|A,W]
estimate W={marginally significant
covariates}
All p-values are FDR adjusted
Simulation Study
> Test methods ability to determine “true”
variables under increasing correlation
conditions
• Ranking by measure and p-value
• Minimal list necessary to get all “true”?
> Variables
• Block Diagonal correlation structure:
10 independent sets of 10
• Multivariate normal distribution
• Constant ρ, variance=1
• ρ={0,0.1,0.2,0.3,…,0.9}
> Outcome
• Main effect linear model
• 10 “true” biomarkers, one variable
from each set of 10
• Equal coefficients
• Noise term with mean=0 sigma=10
–“realistic noise”
THEOREM
• Consider any generalized linear
regression model from the normal,
binomial, Poisson, Gamma, Inverse
Gaussian, with canonical link function, in
which the linear part contains the
treatment variable as main term and
contains intercept. Let r be cts diff. Then
the MLE of r(EY(0),EY(1)) is
asymptotically consistent and locally
efficient.
Median number of false positives
TargetImpact
advantage
Power
Patients
Crude
ANCOVA TargetImpact
Crude
ANCOVA
Viral Load below 50 copies/mL
500
1000
3000
14.6%
31.7%
70.1%
x
x
x
32.3%
60.4%
97.0%
2.4
2.6
2.2
x
x
x
27.5
31.2
30.5
1.4
1.5
1.5
Change in CD4 counts
500
1000
3000
13.1%
23.9%
56.3%
97.5%
100%
100%
99.4%
100%
100%
Average number of patients needed in a GST:
• 2,593 with a crude analysis
• 1,980 with TargetImpact
$15M or 23%
savings (given the
typical cost of $25K per
patient)