Automated Hypothesis Generation Based on Mining Scientific
advertisement
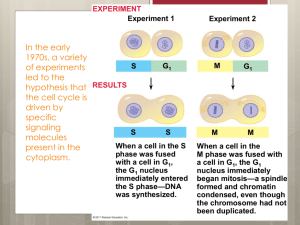
Automated Hypothesis Generation Based on Mining Scientific Literature Scott Spangler, Angela D. Wilkins, Benjamin J. Bachman, Meena Nagarajan, Tajhal Dayaram, Peter Haas, Sam Regenbogen, Curtis R. Pickering, Austin Comer, Jeffrey N. Myers, Ioana Stanoi, Linda Kato, Ana Lelescu, Jacques J. Labrie, Neha Parikh, Andreas Martin Lisewski, Lawrence Donehower, Ying Chen, and Olivier Lichtarge. 2014. Automated hypothesis generation based on mining scientific literature. In Proceedings of the 20th ACM SIGKDD international conference on Knowledge discovery and data mining (KDD '14). ACM, New York, NY, USA, 1877-1886. Kathleen Padova, October 21, 2014 Authorship & Publication • Scott Spangler, Angela D. Wilkins, Benjamin J. Bachman, Meena Nagarajan, Tajhal Dayaram, Peter Haas, Sam Regenbogen, Curtis R. Pickering, Austin Comer, Jeffrey N. Myers, Ioana Stanoi, Linda Kato, Ana Lelescu, Jacques J. Labrie, Neha Parikh, Andreas Martin Lisewski, Lawrence Donehower, Ying Chen, and Olivier Lichtarge • Joint project between Baylor College of Medicine, The University of Texas MD Anderson Cancer Center, and IBM Research • Presented at the 20th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, August 2014 • ~700 downloads from ACM Digital Library Challenge • The amount of information is growing larger than humans’ capability to process. Is there a systematic way we can perform some of this analysis, leaving more time for investigation? Past approaches • Highly structured content where the connections are inferred from the structure (MeSH) • Established empirical laws (chemistry) • Attempts at unstructured content – “hit-ormiss” New approach • KnIT - Knowledge Integration Toolkit o Exploration o Interpretation o Analysis Case Study: p53 Kinases • Protein p53, when chemically modified (phosphorylation) by another protein, is essential in a cell’s own defense against broken, cancerous state • Kinases - proteins that phosphorylate other proteins • Increase in research to find drugs that influence kinases as potential cancer treatments Case Study: Challenge • 500 + kinases X Tens of thousands of possible proteins = Over 10,000,000 possible kinaseprotein combinations • Months to experiment, years to elucidate a single kinase-protein relationship • 33 of 500+ kinases found to modify p53… so far Case study: Challenge • Mine the literature to identify other kinases likely to modify p53 • Create a focused pool of highly likely targets for future experimentation Phase 1: Exploration • • • • • Collect relevant information Design text queries Extract relevant documents (259 kinases) Identify known p53 kinases (23) Model each entity (kinase) Phase 2: Interpretation • Graph the similarity relationships between entities (kinases) • Visualize hidden connections bases on entity models • Discover “deviant” entities based on proximity to other entities sharing similar properties Phase 2: Interpretation Phase 2: Interpretation Phase 3: Analysis • Graphically diffuse annotated similarity relationships throughout graph • Adds a “likeliness” factor to entities • Domain expert verifies the candidates for further study Phase 3: Analysis Computational Validation • Leave-one-out cross-validation o Mark one known p53 kinase at a time as unknown o Can algorithm correctly predict the unknown known? • Retrospective study o Run literature pre-2003 to try to predict the known p53 kinases that would be discovered later • Large scale study o Apply algorithm to different data set to predict kinases that target other (not p53) proteins Experimental Validation Future work • Ramp up capability for larger-scale analysis • Apply to wider area of proteins creating a more comprehensive map of proteins and functions involved in cancer research • Apply the general literature mining approach to other scientific domains Discussion • To what extent is success dependent on partnering outside of the known domain? • How reliable can this technique be evaluating literature over a large timespan where the vocabulary may evolve?