Computational Biology
Dr. Jens Allmer
Lecture Slides Week 3
MBG404 Overview
Processing
Pipelining
Generation
Data
Storage
Mining
Sample preparation for mass spectrometry
Centrifugation of crude
cell extracts in
sucrose gradient
0.5 M
1.3 M
Thylakoids
1.8 M
Starch, etc.
Sucrose
gradient
Separation of the
thylakoid fractions via
SDS PAGE
Cutting of
interesting bands
from the gel
Proteolytic (trypsin)
digestion in gel
Liquid chromatography
of resulting peptides
Mass Spectrometry (MS)
1D SDS PAGE of
thylakoid
fraction from crude cell
extracts
of Chlamydomonas
reinhardtii.
Mass
spectrometric
methods for
protein
identification
Schematic depiction of an ion trap mass spectrometer
Peng, J. and Gygi, S.P. (2001) Proteomics: the move to mixtures. J. Mass Spectrom., 36,
1083-1091.
Example tandem MS spectrum
479.4
100
Scan 4502
626.1
100
90
95
90
+ d Z ms [ 622.30-632.30]
85
85
80
80
75
70
Scan 4501
626.6
2
65
70
65
55
Relative Abundance
50
100
45
40
35
95
30
90
627.1
25
20
85
15
1
80
627.7
10
0
622
623
624
625
626
70
627
m/z
628
629
630
631
Relative Abundance
828.2
55
50
957.3
45
40
30
632
715.2
25
65
958.2
20
60
15
55
+ c Full ms [ 400.00-2000.00]
50
10
835.5
602.2
400
600
0
40
200
982.4
35
30
610.2
1054.4
25
1156.2
852.2
20
503.9578.8
445.1
1157.5
703.2
765.9 885.0
1217.7
1259.8
10
1469.7
5
0
400
600
800
1070.3
406.2
5
45
15
60
35
5
75
3
75
60
626.3
535.8
95
1000
1200
m/z
1400
1600
1800
2000
800
1000
m/z
1200
1400
1600
1800
Mass spectrometric peptide fragmentation spectrum analysis
(Sequest or Mascot)
Ion trap mass spectrometry (ITMS)
Single peptide ions
Mass spectra
Collision-induced dissociation (CID)
Sequentially fragmented
peptide ions
Tandem mass spectra
Database
search
‚In silico‘ tryptic
digestion
Theoretical MS/MS
fragmentation pattern
Peptide
amino acid sequences
‚Hit‘
Significant match with
theoretical fragmentation pattern
of a database sequence
DATABASE
Translated DNAor protein sequences
Cross correlation
• Digesting the database with the enzyme in
question.
• Picking all fragments within a mass window
close to the precursor mass of the peptide in the
mass spectrum
• Calculating an artificial spectrum from all those
fragments
• Cross correlate spectra to original mass
spectrum
WLQYSEVIHAR
Theoretical spectrum in red (a,b,c,x,y,z ions) and measured spectrum in blue
Mass spectrometric peptide fragmentation spectrum analysis
(Sequest or Mascot)
Ion trap mass spectrometry (ITMS)
Single peptide ions
Mass spectra
Collision-induced dissociation (CID)
Sequentially fragmented
peptide ions
Tandem mass spectra
Database
search
‚In silico‘ tryptic
digestion
DATABASE
Translated DNAor protein sequences
Theoretical MS/MS
fragmentation pattern
Peptide
amino acid sequences
Limitation:
‚Hit‘
Significant match with
theoretical fragmentation pattern
of a database sequence
Identification is limited
to peptide sequences present in the database.
Database Search Software
• Many tools have been developed
– OMSSA (NCBI, discontinoued)
– X!Tandem (The global proteome machine)
X!Tandem
• http://www.thegpm.org/tandem/
X!Tandem Initalization Files
• X!Tandem
– Taxonomy.xml
– Default_Input.xml
– Input.xml
• Running X!Tandem
– ?>tandem.exe input.xml
• That was easy
– But behold, what about the input?
OMSSA
• Open Mass Spectrometry Search Algorithm
• Discontinued
– Due to problems?
• Still existing uses
– PeptideShaker
– SearchGUI
Sequence Alignment
• Exact
– simple
target
pattern
• Approximate
– More difficult
target
pattern
Sequence Alignment
• Exact pattern matching
–
–
–
–
Naive method aligns pattern with each location of the target
Boyer-Moore indexes the pattern to skip some alignments
Wu-Manber indexes many patterns and skips some alignments
Indexing
• Suffix tree indexes target and then quickly finds each pattern
• Many other methods
Sequence Alignment
• Approximate pattern matching
– Pairwise
• Local
– Smith Waterman
– BLAST
– FASTA
• Global
– Needlemann Wunsch
– Multiple
• T-Coffee
• ClustalW
• ...
Basic Local Alignment Seach Tool
• Input
– Pattern
– Target
– Search parameters and settings
• Output
– Alignments in various formats
• XML
• Help
– http://www.ncbi.nlm.nih.gov/books/NBK1763/
BLAST
• Target
– Needs to be indexed
– Cannot be FASTA
– Must fit to the pattern and BLAST variant
• protein target and protein pattern can be searched using blastp
• Target indexing
– makeblastdb, in the BLAST package can index FASTA files
– Needs sequence input (e.g. FASTA, asn.1)
– Needs sequence type to be provided e.g.: protein
BLAST
• blastp
– Needs indexed database
– Needs query sequence (can be unindexed FASTA)
– Produces alignments
22
Blast flavors
Query:
DB:
•
•
•
•
•
DNA
DNA
Protein
Protein
BlastN - nt versus nt database
BlastP - protein versus protein database
BlastX - translated nt (6 frames) versus protein database
tBlastN - protein versus translated nt database (6 frames)
tBlastX - translated nt versus translated nt database (both 6 frames)
BLAST Output
• XML
– -outfmt 5
• This switch leads to XML output
End Theory I
• 5 min mindmapping
• 10 min break
Practice I
Download Blast
• http://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE
_TYPE=BlastDocs&DOC_TYPE=Download
– Get blastp and makeblastdb from mbg404 since you are not
allowed to install anything
• Download a Fasta file (protein, genome, collection of
sequences in fasta format)
– Database must consist of amino acids since we only have
access to blastp today
• Use makeblastdb from the Blast package to index the file
• Several files will be created when you do it right
MakeDB
• Example
– makeblastdb -in seq.fasta -dbtype prot -out seqBl –title
seqBlastDB
• More information?
– Go to the doc folder of BLAST
– Documentation is there
– http://www.ncbi.nlm.nih.gov/books/NBK1763/
BLAST
• Now that we have an indexed database try to run BLAST
• Read documentation and try to solve the simplest case
– You will need the indexed database and you will need a FASTA
file as query
– You could create queries from the database and slightly change
them
• Good luck
OMSSA
• Unzip folder and check
– Alternatively, download from NCBI
•
•
•
•
•
MS/MS mgf file
Database file as FASTA
makeblastdb.exe
omssacl.exe
usermods.xml
OMSSA
Before running OMSSA, database file must be converted to
BLAST-like format.
So let’s run makeblastdb.exe to create a hash-indexed
database
OMSSA
Here 2 different settings are used.
First one is with 0.05 product ion tolerance
Second one is with default product ion tolerance
For variable modifications (-mv) check usermods.xml
X!Tandem
• Unzip folder and check
•
•
•
•
Mgf formated spectra (file)
Database file (FASTA)
tandem-win32-10-12-01-1 folder
Used .xml configuration files (default_input.xml, input.xml
and taxonomy.xml)
• To get the same output given in zip folder;
– Replace configuration files in «tandem-win\bin» folder with ones
in «used» folder.
– Also copy database file to «fasta» folder and .mgf file to «bin» in
«tandem-win»
X!Tandem Console Application
X!Tandem Default Input
Parameters such as mass tolerances, enzyme type, number of
charged for search can be reset in default_input.xml
X!Tandem Input.xml
In input.xml file, you should specify path of:
• taxonomy.xml
• default_input.xml
• Spectra filename
• Output filename
NOTE: Here input.xml and all files above are in same folder(directory))
X!Tandem Taxonomy
In taxonomy file, you should specify «database file path». In this
example, database file is in «fasta» folder in «Xtandem\tandem-win3210-12-01-1» folder.
X!Tandem Output
End Practice I
• 15 min break
Theory II
Automation
• BLAST needed sequence file preprocessing
• OMSSA, X!Tandem, etc may need conversion of spectra
files
– A lot of manual processes
• Needed: an automation facility
• Solution: computational pipelines
Computational Pipeline
Spectra
mgf
mzXML
dta
mz2
...
Spectra
Format
Converter
DB
dta
PepNovo
Result
Converter
Analysis Network
Result
Converter
Spectra
mgf
mzXML
dta
mz2
...
2DB
Lutefisk
Spectra
Format
Converter
GPF
PepNovo
Result
Converter
General Pipeline Considerations
• Data cannot be connected to data
• Operations cannot be connected directly either
• Data needs to be transformed (operation)
Data
Store
DB
Data
Store
DB
Data Flow
Operation
Data
X
Operation
• In the example the data element cannot be
directly connected to the DB
• The data element is also not necessar it has
been added to clarify that the process
generates data which will go directly to the DB
Data
OpenMS
Pipeline Examples
• We will see a few examples
– OpenMS/TOPP
– Trans Proteomics Pipeline
– Proteomatics
– Ensembl
TOPP - The OpenMS Proteomics Pipeline
http://open-ms.sourceforge.net/
Trans-Proteomics Pipeline TPP
http://sourceforge.net/projects/sashimi/
Proteomatic
http://www.uni-muenster.de/hippler/proteomatic/
Proteomatic
http://www.uni-muenster.de/hippler/proteomatic/
Ensembl
http://genome.cshlp.org/content/14/5/934.full
http://www.ensembl.org
Standardization
• Some programs have the same aim
– Unfortunately, produce largely different output
– Depend on different input formats
– One need for pipelines arises from this
• Standardization can eleviate that problem
• Currently mostly XML
– Developments of controlled vocabularies are seen
• In ten years full transition to ontologies expected
Standardization (HUPO PSI)
Selfmade
• Windows
– Batch script
– Powershell
• Linux
– Bash script
– Shell script
• Common
– A file that contains instructions
– Usually found in the console
Delete Temp
• Batch script
– cd c:\
– cd Windows\Temp
– rm –r –s *.*
• Save file as
– DeleteTemp.bat
• Put the file into
– C:\Users\%USERNAME%\AppData\Roaming\Microsoft\Windows
\Start Menu\Programs\Startup
• Next startup
– The temporary files will be deleted
Pipelining
• The previous example performed pipelining
• You can use this for anything like
– First making a BLAST DB
– Second searching it
• Advantage
– You have a log of the settings etc.
– You can repeat it at any time
End Theory II
• 5 min mindmapping
• 10 min break
Practice II
Raw Data
• Screenshots
• Copy paste
• Unstructured
• Not integrated
• Unreflected
Information
• Structured Data
PepNovo
PEAKS
Lutefisk
OMSSA
• Sorted
• Integrated
Prediction Distance
1
0.8
0.6
0.4
0.2
0
0.22-0.43
• Properly graphed
• Figure
– Number
– Caption
– Reference
0.55-0.66
0.67-0.83
Spectral Quality
Figure 1: Spectral Quality (present fragment ions/
expected fragment ions) versus Prediction Distance to the
true sequence (normalized edit distance; 0:great and
1:poor). Predictions were done by PepNovo, PEAKS, and
Lutefisk while identification was done with OMSSA. All
MS/MS spectra were of charge 1.
Even Better
PepNovo
COMAS
Lutefisk
OMSSA
PEAKS
1
Prediction Quality
0.8
0.6
0.4
0.2
0
0.22-0.43
0.55-0.66
0.67-0.83
Spectral Quality
Figure 5: Spectral Quality (present fragment ions / expected fragment ions)
versus Prediction Quality (normalized edit distance). The box-and-whisker
plot presents three groups at different spectral quality. Note there were no
measurements between 0.43 and 0.55, before 0.22 and after 0.83.
Presenting Data
• When presenting data in your manuscripts:
• Raw data (not acceptable)
• Information (minimum)
• Knowledge (strive for this)
Whiteboardmaths.com
Stand SW 100
© 2004 - 2008 All rights reserved
Click when ready
In addition to the demos/free presentations in this area there are at
least 8 complete (and FREE) presentations waiting for download under
the My Account button.
Simply register to download immediately. 63
www.similima.com
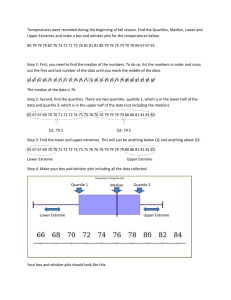
Median, Quartiles, Inter-Quartile Range and Box Plots.
Measures of Spread
Remember: The range is the measure of spread
that goes with the mean.
Example 1. Two dice were thrown 10 times and their
scores were added together and recorded. Find the mean
and range for this data.
7, 5, 2, 7, 6, 12, 10, 4, 8, 9
Mean = 7 + 5 + 2 + 7 + 6 + 12 + 10 + 4 + 8 + 9
10
= 70
=7
10
Range = 12 – 2 = 10
www.similima.com
64
Median, Quartiles, Inter-Quartile Range and Box Plots.
Measures of Spread
The range is not a good measure of spread because one
extreme, (very high or very low value) can have a big
affect. The measure of spread that goes with the
median is called the inter-quartile range and is
generally a better measure of spread because it is not
affected by extreme values.
A reminder about
the median
www.similima.com
65
Averages (The Median)
The median is the middle value of a set of data once
the data has been ordered.
Example 1. Robert hit 11 balls at Grimsby driving
range. The recorded distances of his drives, measured
in yards, are given below. Find the median distance for
his drives.
85, 125, 130, 65, 100, 70, 75, 50, 140, 95, 70
50, 65, 70, 70, 75, 85, 95, 100, 125, 130, 140
Single middle value
Ordered data
Median drive = 85 yards
www.similima.com
66
Averages (The Median)
The median is the middle value of a set of data once
the data has been ordered.
Example 1. Robert hit 12 balls at Grimsby driving range.
The recorded distances of his drives, measured in yards,
are given below. Find the median distance for his drives.
85, 125, 130, 65, 100, 70, 75, 50, 140, 135, 95, 70
50, 65, 70, 70, 75, 85, 95, 100, 125, 130, 135, 140
Two middle values so
take the mean.
Ordered data
Median drive = 90 yards
www.similima.com
67
Finding the median, quartiles and inter-quartile range.
Example 1: Find the median and quartiles for the data below.
12,
6,
4,
9,
8,
4,
9,
8,
5,
9,
8,
10
10,
12
Order the data
Q2
Q1
4,
4,
5,
6,
Lower
Quartile
= 5½
8,
8,
Q3
8,
Median
= 8
9,
9,
9,
Upper
Quartile
= 9
Inter-Quartile Range = 9 - 5½ = 3½
www.similima.com
68
Finding the median, quartiles and inter-quartile range.
Example 2: Find the median and quartiles for the data below.
6,
3,
9,
8,
4,
10,
8,
4,
15,
8,
10
Order the data
Q2
Q1
3,
4,
4,
6,
Lower
Quartile
= 4
8,
8,
Median
= 8
Q3
8,
9,
10,
10,
15,
Upper
Quartile
= 10
Inter-Quartile Range = 10 - 4 = 6
www.similima.com
69
Discuss the calculations below.
Battery Life:
The life of 12 batteries recorded in hours is:
2, 5, 6, 6, 7, 8, 8, 8, 9, 9, 10, 15
Mean = 93/12 = 7.75 hours and the range = 15 – 2 = 13 hours.
2, 5, 6, 6, 7, 8, 8, 8, 9, 9, 10, 15
Median = 8 hours and the inter-quartile range = 9 – 6 = 3 hours.
The averages are similar but the measures of spread are
significantly different since the extreme values of 2 and 15 are
not included in the inter-quartile range.
www.similima.com
70
Box and Whisker Diagrams.
Box plots are useful for comparing two or more sets of data like
that shown below for heights of boys and girls in a class.
Anatomy of a Box and Whisker Diagram.
Lower
Lowest
Quartile
Value
Whisker
4
5
Median
Upper
Quartile
Whisker
Box
6
7
Highest
Value
8
9
10
11
12
Boys
130
140
150
160
www.similima.com
170
180
cm
190
Girls
Box Plots
71
Drawing a Box Plot.
Example 1: Draw a Box plot for the data below
Q2
Q1
4,
4,
5,
6,
8,
8,
Lower
Quartile
= 5½
4
5
Q3
8,
Median
= 8
6
7
8
www.similima.com
9
9,
9,
9,
10,
12
Upper
Quartile
= 9
10 11
12
72
Drawing a Box Plot.
Example 2: Draw a Box plot for the data below
Q2
Q1
3,
4,
4,
6,
8,
Lower
Quartile
= 4
3
4
5
6
Q3
8,
8,
Median
= 8
7
8
9
www.similima.com
9,
10,
10,
15,
Upper
Quartile
= 10
10 11
12 13
14 15
73
Drawing a Box Plot.
Question: Stuart recorded the heights in cm of boys in his
class as shown below. Draw a box plot for this data.
Q2
QL
Qu
137, 148, 155, 158, 165, 166, 166, 171, 171, 173, 175, 180, 184, 186, 186
Lower
Quartile
= 158
130
140
Upper
Quartile
= 180
Median
= 171
150
160
www.similima.com
170
180
cm
190
74
Drawing a Box Plot.
Question: Gemma recorded the heights in cm of girls in the same class and
constructed a box plot from the data. The box plots for both boys and girls
are shown below. Use the box plots to choose some correct statements
comparing heights of boys and girls in the class. Justify your answers.
Boys
130
140
150
160
170
180
cm
Girls
1. The girls are taller on average.
2. The boys are taller on average.
3. The girls show less variability in height.
5. The smallest person is a girl.
www.similima.com
4. The boys show less variability
in height.
75
6. The tallest person is a boy.
190
Konstanz Information Miner
• We will use the Workflow Management and Data
Analytics Platform
• First we need to find out how to get our data into
KNIME
Create Data
• Use Excel to create two colums
– Girls, boys
• Make a few hundred random numbers (randbetween)
– 140 -170 for girls
– 150 - 180 for boys
• Copy the table
• Paste into Notepad++
• Save as Distribution.txt
KNIME Data Import
• Open Knime
• Select the folder containing the data as workspace
• Right click LOCAL
– Select new workflow
– Name it HeightAnalysis
• Drag and Drop Distribution.txt into the workflow
Box Plot
• Type box to find box plot
node
• Double click
• Right click Box Plot node
– Select Execute and open
views
• Done
Workflow