Document
advertisement

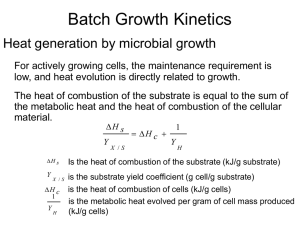
Exponential phase = log-phase Maximum growth rates μmax „midexponential“: bacteria often used for functional studies Max growth rate -> smallest doubling time Vmax S v K M S • Used when microbe population is constant = non-growing (or short time spans) • Derivable from first principles (enzyme-substrate binding rates and equilibria expressions) • Parameter determination methods used for Monod calculations (i.e. Lineweaver Burke) •Relates specific growth rate, , to substrate concentration •Empirical---no theoretical basis—it just “fits”! •Have to determine max and Ks in the lab •Each is determined for a different starting S max S Ks S Michaelis Menten Kinetic expression derived (theoretical) Constant enzyme pool Monod Empirical expression Growth Free enzymes Non-growing microbes v vs. S where v is velocity Km is half saturation constant Enzyme concentration increases with time Relates microbial growth rate constant to S μ vs S Ks is half saturation constant Parameters (vmax or μmax; Ks or Km) are determined by linearization (e.g. Lineweaver Burke model) or nonlinear curve fitting. Relationship between dependent variable and S determined experimentally, in the lab Range of S Set conditions (T, chemistry, enzyme or microbe) Measure the v or μ for each S Plot v or μ vs. S; analyze data for parameter estimation • Double reciprocal plot (Lineweaver Burke) • Commonly used • Caution that data spread are often insufficient • Other linearization (Eadie Hofstee) • Less used, better data spread • Non-linear curve fitting • More computationally intensive • Progress-curve analysis (for substrate depletion) • Less lab work (1 curve), more uncertainty It applies where μ ǂ 0 -> exponential growth (μ = μmax ) + transition into stationary max S Ks S KS is the half-saturation coefficient [mg/L] Monod kinetics -> “Substrate depletion kinetics” Since And Then And dX dS X Y dt dt max S Ks S Y Yield coefficients Monod applies!! dS X max SX dt Y (K s S)Y max Where k = dS kSX Y dt K s S k is the maximum substrate utilization rate [sec-1] KS is the half-saturation coefficient [mg/L] Substrate consumption rates have often been described using ‘Monod kinetics’ -> Substrate controls growth Kinetics dS kSX dt K s S S is the substrate concentration [mg/L] X is the biomass concentration [mg/ L] k is the maximum substrate utilization rate [sec-1] KS is the half-saturation coefficient [mg/L] Stoichiometric Coefficients for Growth Yield coefficients, Y, are defined based on the amount of consumption of another material. X dX dt YX / s ds s dt Because ΔS changes with growth condition, YX/S is not a constant S << KS 3 mixed order 2 S >> KS 1 max , 1/hr Stationary phase μ=0 S, mg/L Expontential growth μ = μmax 1. Zero-order region, S >> KS, the equation can be approximated by μ = μmax -> exponential growth dS kX dt S << KS 2. Center region, Monod “mixed order” kinetics must be used -> transition from exponential growth to stationary growth caused by [S] limitation 3 2 S >> KS 1 max dS kSX dt K s S 3. First-order region, S << KS, the equation can be approximated as μ = μmaxS/Ks -> transition from exponential growth to stationary growth caused by [S] limitation Just before stationary phase starts (stationary phase μ = 0) mixed order , 1/hr dS kSX dt Ks k is the maximum substrate utilization rate [sec-1] KS is the half-saturation coefficient [mg/L] dS kSX dt Ks S, mg/L dS kX dt Three common assumptions Monod kinetics applies (mid range concentrations) -> “Substrate depletion kinetics” First-order decay (low concentration of S, applicable to many natural systems) Zero-order decay (substrate saturated) μ =μmax -> exponential growth Cellular growth rate Monod approximation dX μ X dt dX s X μmax dt Ks s s μ μmax Κs s Yield factor X dX dt YX / s ds s dt Substrate Utilization YP / X P dP dt dX X dt ds μmax s X dt YX / s K s s Product Formation (Beginning of Stationary Phase) dP s X YP / X max dt Ks s Rate per microbe, which depends on Species Substrates Environmental factors Total numbers of microbes • Culture-based (limited: 2000 species vs. 13,000 species of bacteria in soil by DNA-based methods •Counting colony forming units (CFUs) •Activity assays: need cell or biomass count to normalize • Culture-independent •Direct Counts •General fluorescent stain, like acridine orange or SYBR gold •Counting cells in FISH assay •Biomass assays •Quantification of an element like C or N •Chloroform fumigation / incubation or direct extraction •Total protein or DNA -> Why is it important to know the kinetics of the reaction in the fermenter? -> What is going on in a fermenter? -> How to control the process in a fermenter? Stochiometric Coefficients -> Too complex !!!! -> Blackbox effect substrates + cells → extracellular products + more cells ( ∑S + X → ∑P + nX) Monod’s model -> S depletion 1. Mass balance : depentend on reactor type -> S, P, X 2. Growth Kinetics: -> Monod model (substrate depleting model) -> Describes what happens in the reactor in steady state (constant conditions) Primary metabolic products Secondary metabolic products Microbial Products 1. Growth associated products : products appear simultaneoulsy with cells in culture 1 dP qp YP / X X dt qp is the specific rate of product formation (mg product per g biomas per hours 2. Non-growth associated products : products appear during stationary phase of batch growth q p cons tan t 3. Mixed-growth associated products : products appear during slow growth and stationary phase q p Biotechnological processes of growing microorganisms in a bioreactor Mass Balance: Fin = Fout = 0 V= const. Fin ≠ 0; Fout = 0 V increases Fin = Fout ≠ 0 V = const. 1. Mass balance : depentend on reactor type -> S, P, X 2. Growth Kinetics: -> Monod model (substrate depleting model) -> Describes what happens in the reactor in steady state (constant conditions) 1. Mass Ballance: Biomass: In – Out + Reaction = Accumulation FX0 - FX + ∫r dV = dn/dt r = dX/dt = µ X 2. Monod Kinetics: max S Ks S dn/dt = d(XV)/dt dn/dt=V (dX/dt) + X (dV/dt) 3. Steady state: dX/dt = 0 (NOT for Batch reactor!!!) Closed Well-mixed Constant volume -> substrate growth limiting factor V= const. Mass Balance - Biomass: Acc = dn/dt Verbal: n = mole In – Out + Reaction = Accumulation dn/dt = d(XV)/dt = (dX/dt) V Math: 0 0 rV dX/dt V Rearrange: r V = dX/dt V Growth -> Substrate concentration controls growth rate dX μ X r dt Growth Cellular growth rate Monod approximation dX μ X dt dX s X μmax dt Ks s s μ μmax Κs s Yield factor X dX dt YX / s ds s dt Substrate Utilization YP / X P dP dt dX X dt ds μmax s X dt YX / s K s s Product Formation (Beginning of Stationary Phase) dP s X YP / X max dt Ks s Biotechnological processes of growing microorganisms in a bioreactor Mass Balance: Fin = Fout = 0 V= const. Fin ≠ 0; Fout = 0 V increases Fin = Fout ≠ 0 V = const.