Amino Acids and Peptides
advertisement

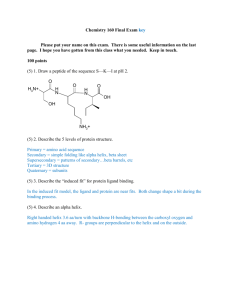
Amino Acids: Structure, Analysis, and Sequence (in peptides) Structures of the Amino Acids O C H2N C OH O H H2N C C OH O H HO C C general structure of an amino acid L-serine O C H2N H C HO L-glyceraldehyde H H CH2OH CH2OH CH2OH R O H Fischer projection of L-glyceraldehyde OH H Fischer projection of an L-amino acid R Examples of the 20 common amino acids: Neutral amino acids: -R = -H, -CH3, -CH2CH(CH3)2, -CH2SH, -CH2OH, -CH2C6H5, -CH2C6H4OH Acidic amino acids: -R = -CH2CO2H, -CH2CH2CO2H Basic amino acids: -R = -CH2CH2CH2CH2NH2, -CH2CH2CH2NHC(NH2)2 Abbreviations of Amino Acids • Amino acids have 1-letter and 3-letter abbreviations; the 1-letter abbreviations are used almost exclusively today, but you should also be aware of the older 3-letter abbreviations. • Some examples: – – – – – – – – glycine (R = H) alanine (R = CH3) phenylalanine (R = CH2C6H5) tyrosine (R = CH2C6H4OH) serine (R = CH2OH) cysteine (R = CH2SH) methionine (R = CH2CH2SCH3) leucine (R = CH2CH(CH3)2) Gly Ala Phe Tyr Ser Cys Met Leu G A F Y S C M L Isoelectric Point • Each amino acid has an isoelectric point, (pI) numerically equal to the pH at which the zwitterion concentration is at a maximum. • The amino acid has no NET charge at its pI; it has one positive and one negative charge. • At a pH less than the value of the isoelectric point, the amino acid is protonated and has a POSITIVE charge; at a pH greater than the pI the amino acid is deprotonated and has a NEGATIVE charge. O C H3N O OH C H H R H3N O O C OH H R O H2N H R @ pH < pI @ pH = pI @ pH > pI Cation Neutral Anion (zwitterion form) Separation and Analysis using pI values • Differences in isoelectric points (and therefore charges) are used to separate mixtures of amino acids by two common methods: – Ion exchange chromatography – Polyacrylamide gel electrophoresis (PAGE) These methods will be illustrated with a simple mixture of three amino acids having very different isoelectric points: H O H3N C CO Mixture of: buffered at pH 6.0 CH2CO2 H O H O + D (pI=2.8) aspartic acid H3N C CO + CH3 A (pI=6.0) alanine H3 N C CO CH2CH2CH2CH2NH3 K (pI= 9.7) lysine Ion Exchange Chromatography H O H3N C CO Mixture of: buffered at pH 6.0 CH2CO2 + H3N C CO D (pI=2.8) SO3 K sulfonated polystyrene H O H O SO3 + H3 N C CO CH2CH2CH2CH2NH3 CH3 A (pI=6.0) K (pI= 9.7) (strongly retained) A (slightly retained, & D (unretained) SO3 D- elutes first, followed by A; K+ elutes last, and only after pH of buffer is increased and K+ is deprotonated. Ion Exchange Chromatography • Recall that in our simple mixture D- elutes first, followed by A; K+ elutes last, and only after the pH of buffer is increased and K+ is deprotonated. • But there is a problem in detecting amino acids; they are colorless, and most of them have very little absorption in the UV region (they have no conjugation, except in the four aromatic amino acids) • To overcome this difficulty, amino acids are converted (after separation by ion exchange chromatography) to a derivative using ninhydrin. Derivatization with Ninhydrin O O H O OH 2 + OH O N H3N C CO O R(any) O O Ninhydrin (2 mol) reacts with one mol of ANY amino acid to give the SAME blue colored product. This reaction is performed post-column, after Ion Exchange Chromatography separation of a mixture of amino acids. The area of each peak in the chromatogram is proportional to the relative molar amount of the amino acid of that retention time. Ion Exchange Chromatography Recall that in our simple mixture D- elutes first, followed by A; K elutes last, and only after the pH of buffer is increased and K+ is deprotonated. D injection A Increase pH of buffer Retention time K Polyacrylamide Gel Electrophoresis (PAGE) H O H3N C CO Mixture of: buffered at pH 6.0 CH2CO2 H O H O + H3N C CO H3 N C CO + CH2CH2CH2CH2NH3 CH3 D (pI=2.8) A (pI=6.0) Before current is turned on: K A D After current is turned on: K A D K (pI= 9.7) The Strecker amino acid synthesis O CH3CH NH NH3 KCN, H2O NH2 CH3CH CN CH3CH H3O heat + NH2 CH3CH CO2H (racemic alanine) Resolution of racemic amino acids CO2H D- H NH2 H O O R CH3COCCH3 + NHCCH3 H NHCCH3 R R Carboxypeptidase + CO2H L- H2N CO2H O CO2H O H R Racemic amino acid O + CO2H CH3CHN H R Racemic N-acetyl amino acid CO2H H2N H R L-amino acid + D-N-acetylamino acid Carboxypeptidase hydrolyzes the amide bond ONLY of the L-aa, leaving the unnatural D-N-acetylamino acid unreacted; separation is simple Covalent bonding in peptides • Amino acids are covalently bonded to one another by amide linkages (bonds) between the carboxylic acid group of one amino acid and the amino group of the next amino acid. • Amide bonds are strong and are resistant to hydrolysis, but there are enzymes that catalyze their hydrolysis (to the amino acids). peptidase enzyme H O H2N C C R1 H H O N C C OH R2 peptidase enzyme H O H2N C C OH R1 H O + H2N C C OH R2 • In addition to amide bonds, a second kind of covalent bond exists in some peptides in which two cysteine residues (amino acid units) are connected through a disulfide bond formed by oxidation (dehydrogenation) of the sulfhydryl (SH, thiol) groups (next slide). peptidase enzyme Disulfide bonding in peptides H H O H H O H H O H H O H H O H H O N C C N C C C C C CH3 C N CH2 C N CH3 C N C N CH(CH3)2 H C CH2CH2SCH3 SH SH CH2 C H C N C O H H O H C CH(CH3)2 CH3 N C C H O H N C C H O H CH2OH N C CH3 C N C C N H O H H O H H [O] H H O H H O H H O H H O H H O H H O N C C N C C C C C CH3 C N CH2 C N CH3 C N C N CH(CH3)2 H C CH2CH2SCH3 S S CH2 C H C N C O H H O H C CH(CH3)2 CH3 N C C H O H N C C H O H CH2OH N C CH3 C N C C N H O H H O H H Total Hydrolysis: conversion of a peptide into a mixture of its component amino acids H O H H O H H3N C C N C CH3 C N CH2OH A H O H H O H H O H H O C C C C C N CH2 C N CH(CH3)2 H F S C N CH2CH2SCH3 M G V C O H3O, heat (total hydrolysis) H O H3N C CO CH3 A H O + H3N C CO CH2OH S H O H O + H3N C CO CH2 F + H3N C CO H O H O + H3 N C CO CH(CH3)2 + H3N C CO CH2CH2SCH3 H V G M (equimolar mixture of A, S, F, V, G, and M ) S Ion Exchange Chromatogram: G A V M F 2. Amino Acid Sequence: Primary Structure Determination of Peptides • Total hydrolysis followed by and ion exchange chromatography and then ninhydrin derivatization tells us the identity and relative amount of each amino acid present in the peptide • It gives NO INFORMATION about the sequence, or order of attachment of the amino acids, however. • For this, we need to perform selective hydrolysis of the peptide. • We’ll learn three methods: – Sanger’s reagent followed by total hydrolysis – Carboxypeptidase – Leucine aminopeptidase Sanger’s Reagent: N-terminal Amino Acid Analysis H O H H O H H O H H O H H O H H3N C C N C C N CH3 A CH2OH S H O C C N C C N C C N C C O CH2 CH(CH3)2 H CH2CH2SCH3 F V M G Sanger's Reagent (2,4-dinitrofluorobenzene) O2N H H O H H O H H O H H O H H O H H O N C C N C C N CH3 NO2 CH2OH C C N C C N C C N C C OH CH2 CH(CH3)2 H CH2CH2SCH3 Sanger’s Reagent, cont’d O 2N H H O H H O H H O H H O H H O H H O N C C N C C C N C C C CH2 CH(CH3)2 H CH3 C N CH2OH C N C N C OH CH2CH2SCH3 NO2 H3O, heat (total hydrolysis) H H O O2N N C C OH + CH3 "tagged" A H O H3N C CO CH2OH S H3N C CO CH2 F + H3N C CO H O H O H O H O + NO2 + H3N C CO + H3N C CO CH(CH3)2 H CH2CH2SCH3 V G M ("tagged" A plus an equimolar mixture of S, F, V, G, and M ) Carboxypeptidase: C-terminal AA Analysis H O H H O H H3N C C N C CH3 C N CH2OH A H O H H O H H O H H O C C C C C N CH2 C N CH(CH3)2 H F S C N CH2CH2SCH3 M G V C O Carboxypeptidase H O H H O H H3N C C N C CH3 C N CH2OH H O H H O H H O C C C C N CH2 C N H O CO H3N C CO + CH(CH3)2 H CH2CH2SCH3 M Carboxypeptidase H O H H O H H O H H O H3N C C N C C N C C N C CH2 CH(CH3)2 CH3 CH2OH CO H O + H3 N C CO H G Ion Exchange Chromatograms following Carboxypeptidase 10 min S G A V M F S G A V M F 20 min 30 min 40 min Leucine aminopeptidase: N-terminal AA Analysis H O H H O H H3N C C N C CH3 C N CH2OH A H O H H O H H O H H O C C N C C C N C CH2 CH(CH3)2 H C N F S C O CH2CH2SCH3 M G V Leucineaminopeptidase, 10 min H O H H O H3N C CO + H3 N C C N CH2OH CH3 A (first aa released) H O H H O H H O H H O C C C N C C C O C N CH2 CH(CH3)2 H F S C N CH2CH2SCH3 G V M Leucineaminopeptidase, 10 more min H O H3 N C CO CH2OH S (2nd aa released) + H3N H O H H O H H O H H O C C N C C C CH2 CH(CH3)2 H F C N V C N C O CH2CH2SCH3 G M Ion Exchange Chromatograms following Leucine Aminopeptidase 10 min S G A V M F S G A V M F 20 min 30 min 40 min Partial Hydrolysis H O H H O H H3N C C N C CH3 C N CH2OH A H O H H O H H O H H O C C C C C N CH2 C N CH(CH3)2 H F S C N V C O CH2CH2SCH3 G M Peptide represented schematically: A S F V G M + dil H3O A S A A S F F V + + V G M + S F V + + G M G M (some molecules) (other molecules) (some other molecules) (different molecules of the peptide can fragment differently, leading to a mixture) Putting it all together! • Suppose an unknown hexapeptide gave “tagged” A (alanine) upon treatment with Sanger’s reagent, and upon treatment with carboxypeptidase, the first amino acid released was M (methionine) followed by G (glycine). Where are M and G? • Partial hydrolysis gave the following identifiable tripeptides: V-G-M, A-S-F, and S-F-V. What is the 1º structure of the hexapeptide (written as usual, with the N-terminal aa on the left and A S F V G M the C-terminal aa on the right)? A S F V G M V G M A S F V G M N-terminal C-terminal