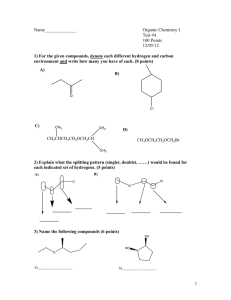
a medicinal chemists view of diversity - UK-QSAR

medicinal chemistry design challenges
chemically intelligent data mining multiparameter optimisation for medicinal chemists how to handle petabytes of data – Google Chemistry!
activity prediction
Dr Tony Wood
VP, Head of Worldwide Medicinal Chemistry
Pfizer Global Research and Development anthony.wood@pfizer.com
the challenge for design?
2002-2005 the primary causes of attrition were safety and pharmacology
in vivo toxicity
results of an analysis of 349 studies on 315 compounds covering 90 targets at 985 doses with >10,000 organ evaluations in 4 species
PK known for all cases with strong correlation between AUC and Cmax compound set has similar diversity to Pfizer file
High Concentration
Toxic Exposures
CmaxLowTox
Uncertain
CmaxHiCln
Minimum exposure with observed toxicity
(CmaxLowTox). Set to an arbitrarily high number if no toxic event is observed at any dose
Maximum exposure without observed toxicity (CmaxHiCln). Set to zero if toxicity was observed at all doses assessed
Clean Exposures
Low Concentration
toxicity threshold selection
300
250
200
150
100
50
0
100nM 1uM 10uM 100uM 1000uM
Threshold (Total Drug) exposure thresholds were chosen to obtain a balance of toxicity/non-toxicity. set to 10uM for the total-drug threshold.
approx 40% of evaluations above threshold & 40% below.
Clean
Uncertain
Toxic similar analysis for free drug levels gives a threshold of 1 uM.
TPSA and clogP are key
the y-axis here is a generalized odds, i.e., the ratio of the probability of a compound with a given parameter value being toxic to the probability of it not being toxic
toxicity odds
combining low TPSA and high cLogP exacerbates the risk
(numbers in parentheses indicate number of outcomes in database) holds for both free-drug or total-drug thresholds ratio of toxic to non-toxic outcomes
toxicity and promiscuity
ratio of promiscuous to nonpromiscuous compounds
TPSA>75 TPSA<75
ClogP<3 0.25 (25) 0.80 (18)
ClogP>3 0.44 (13) 6.25 (29) promiscuity defined as >50% activity in >2 Bioprint assay out of a set of 48 (selected for data coverage only)
ClogP > 3
clogP and organ toxicity
does a good cell viability profile increase the probability of a compound being a CNS CAN w/o organ tox in the clogp risky group (clogp>3)?
15%
20 23
25%
39% 39%
THLE Cv bin x < 25 uM
25 < x < 100 uM x > 100 uM
60%
2 22
22%
5% 14%
ClogP < 3 50% 50% organ tox or not attrition CNS
CANs set
82%
No Organ Tox Organ Tox
DEREK
“a place to store toxicological knowledge” knowledge-based expert system broad range of toxicity endpoints covered identifies structural alert provides literature-based rationale for prediction qualitative or semi-quantitative predictions now has an API for integration into 3 rd party software products
what’s in DEREK?
main strengths are mutagenicity, chromosome damage, carcinogenicity and skin sensitization some recent efforts in hepatotoxicity and teratogenicity
100
90
80
70
60
50
40
30
20
10
0
Endpoint
challenge #1
these relationships were determined using a small well characterised data set much more data lies in non curated data sets with no structure keys we need chemically intelligent data mining to derive knowledge including SAR from this resource
properties of CNS drugs
90%
≥ 0.36
lipE CLOGP 95% Range LOGD_7.4
LE
Drugs
CANs
.2
.3
.4 .5
.6
.7
.8
.9
1 1.1 1.2
-1 0 1 2 3 4 5 6 7 8 9 10 11 12 -7 -6 -5 -4 -3 -2 -1 0 1 2 3 4 5 6 7 8
Median
Drugs: 0.52
CANs: 0.47
LLE
6.2
6.3
ClogP
2.9
3.4
-7 -6 -5 -4 -3 -2 -1 0 1 2 3 4 5 6 7 8
ClogD
1.8
2.3
TPSA MW HBDON BASIC1PKA
Drugs
CANs
0 100
Median
Drugs: 47
CANs: 53
200 100 200 300 400 500 600 700
MW
305
360
0 1 2 3 4 5 6
HBD
1.0
1.0
1 2 3 4 5 6 7 8 9 10 11 12 13 pKa
8.4
8.4
CNS MPO summary
for design (prospective, accurate and constant) increasing CNS MPO enhances the probability of candidate survival and alignment of in-vitro
ClogP
TPSA
C,P,S
P,S
P ermeability
(including efflux)
1
0.8
0.6
0.4
0.2
0
-2 -1 0 1 cLogP
2 3 4 5 6
1
0.8
0.6
0.4
0.2
0
0 20 40
PSA
60 80 100 120
↑
CNS MPO increases the probability of successfully aligning attributes
ClogD
MW
C,S
P
C learance
1
0.8
0.6
0.4
0.2
0
-2 -1 0 1
LOGD7.4
2 3 4 5
1
0.8
0.6
0.4
0.2
0
100 200 300
MW
400 500 600
CNS MPO
Desirability
124
HBD
P
S afety
(including high risk space)
1
0.8
0.6
0.4
0.2
0
0 1 2 3
HBDONORCNT
4 5
26%
56%
75
44% pKa
P,S
1
0.8
0.6
0.4
0.2
0
2 4 6 pKa (1)
8 10
74%
83%
12
17%
71%
34
29%
Drugs CANs
D rug s CAN Pre CAN
Binned Binned Drug_or_CAN_Class (2)
Le a d s
challenge #2
design is now based on a probabilistic basis using complex MPO relationships we need transparent easy to construct and understand methods to perform multiparameter optimisation
chemoinformatic predictions
Pfizer in house
Inpharmatica
StARLITe
Serine proteases unified db
4.8 M structures
275k active compounds
600k activities (IC50, etc)
3k targets
800 human targets
Cysteine proteases
Kinases
Aspartyl proteases
Phosphodiesterases
Metalloproteases
Ion Channels
GPCRs (others: classes A, B & C)
Aminergic GPCRs
Peptide GPCRs
Enzymes
(hydrolases, transferases, oxidoreductases & others)
Nuclear hormone receptors Miscellaneous
Cerep
BioPrint
Thomson
IDDB node : target edge : compound
Bayesian learning
data set (assay data)
“good” actives
“bad” inactives fingerprint bits ~ substructures
Rev Thomas Bayes ca 1702 - 1761 fingerprints are calculated for each molecule check how often fingerprint bit is observed and how often in “good” compound assign weighting factor taking into account both activity ratio and sampling size for instance: “good”/total ratio of
90/100 is statistically more relevant than 9/10 model distinguishes “good” from “bad” predict likelihood molecule is “good
Bayesian model
mining large data sets (HTS)
confirmed measurement
0.45
0.40
0.35
HTA+ > HTA false positive HTA+ or false negative HTA predictions all false negative HTA colored by Bayesian score red: high confidence blue: low confidence
0.30
0.25
0.20
0.15
HTA > HTA+: false positive HTA or false Negative HTA+ predictions red: false HTA+ negative blue: false HTA positive
0.10
0.05
0.05
0.10
0.15
0.20
0.25
0.30
0.35
0.40
0.45
LE HTA
238k actives
( 10 M) human target
mw < 1000
pass reactivity filter
10 actives / target
90% / 214k
predicting promiscuity
3870 compounds with
10,806 predictions
FCFP_6
698 models
Bayesian score
searching virtual space
BIG LEAP: searching the Pfizer liquid and virtual compound collections real: 0.000025%
1.2M singletons derive Bayesian model that distinguishes library
1 from 2, from 3, etc
Pfizer global virtual library
~ 10 12 compounds
5000 libraries
2.5M compounds liquid screening file predict 16 libraries to which compound could belong search only these libraries, in real and virtual compound space
BIG LEAP
Acids
A1
O
B1
N
N
Amines
O x
?
x x
1
Cl
A2
N
O
N
O
O
CF
3 o 1
N
B2
N N
2
*
B4 model is built from synthesized compounds (yellow squares) nearly all fingerprint features of any virtual compound (square marked with “?”) are shared by at least one compound from the training set (squares marked with “X”) virtual products in areas 1 share at least one monomer with a compound from the training set-for compound “O”, the new monomer B2 is very close to previously used B1 compounds from area 2 can be considered outside the scope of the model because they have few fingerprint features in common with the existing products as shown for compound “*” where monomers A2 and B4 are unlike previously used monomers
a new series for PRA
acidic ex-PR library ex-PR library ex-PR library ex-PR library new
CCT services
a framework for computational scientists to publish services (protocols, models) that can be immediately leveraged by project teams a knowledge repository for Computational Scientists to capture and share their best practices when protocols are published they are automatically wrapped as new
PLP component
ligand idea generators
uncharted chemical space
challenge #3
we are not short of idea generators!
easy to construct vast virtual libraries we need ways of rapidly scoring and searching petabytes of data
HERG binding model
training set 98,155 compounds (80%) talidation set 19,577 compounds (20%) test set 9,241 compounds training: Kappa: 0.61
, Concordance 80% training: Sensitivity 81% , Selectivity 80%
2000 test: Kappa: 0.46
, Concordance 74% test: Sensitivity 75% , Selectivity 74%
1500
“Grey zone”, uncertain prediction
>60%
1000
>70% >85%
500
>85%
>95% >95%
0
-80 -70 -60 -50 -40 -30 -20 -10 0 10 20 30 40
No Dofetilide Dofetilide
Inactives
Actives prediction is checked against activity of at least
3 nearest neighbours to generate additional confidence measure
HLM stability model
statistical fingerprint-based model (FCFP-6, Scitegic) unstable well predicted, stable not
Unstable
HLM stability experiment:
Stable
Moderately stable
Unstable Experiment
Stable
Stable
Prediction
Unstable
V1a: design for stability
short t1/2
100
Series 1
Series 2
80
Stable but likely to be poorly absorbed orally
40 long t1/2
20
0
-1.5
-1 -0.5
0 0.5
1 1.5
2 2.5
CLOGP most compounds stable within this cLogP range synthetic effort weighted by desirability
LiPE and LE are quality indicators
linker replacement
O
N N aryl switch
N
V1a Ki 28 nM
MW 441 cLogP 5.5
t1/2 HLM 6 mins
(Human Liver Microsomes)
N
O side chain deletion/replacement
LE: 0.31
LiPE: 1.9
N
N
N
N N
N needed
V1a Ki 780 nM
MW 334 cLogP 1.0
t1/2 HLM 120 mins
LE: 0.32
LiPE: 4.8
LE (Ligand Efficiency) =
-1.4 log (IC50) n Heavy Atoms
“how efficient each heavy atom is”
Class dependent…0.3 – 0.5
LiPE (Lipophilic Efficiency) = -log (IC50) - cLogP
“how efficient each lipophilic fragment is”
using LiPE to view SAR
LiPE= -log (IC50) - cLogP pIC50
8
10nM
7
100nM
LiPE=6
1
6
M
5
1
LiPE=5
LiPE=4
LiPE=3
LiPE=2
2 3 4 5 cLogP
predicting activity
-10 -8
D
G(expt)
-6 -4 -2 0
0
-10
-20
-30 plot of experimental affinity versus calculated enthalpy for reference:
2 kcals = 26-fold off
4.2 kcals = 1000-fold off
-40 kcal/mol
"Improving Accuracy in Protein-Ligand Affinity Calculations"
Paper #104, ACS meeting in Philadelphia (Aug 2004)
Michael K. Gilson, Center for Advanced Research in Biotechnology,
Rockville, MD
the source of the problem?
D
G o D D o
U W T S config
-5 to -10 kcals 15 to -25 kcal 15 to -25 kcal we usually focus on the interactions D <U+W> potential energy force field (CHARMM, AMBER, etc.) van der Waals
Coulombic
Hydrogen-bonding solvation surface area term:
Hydrophobicity/organophilicity we always neglect T D S config generalized Born/Poisson-Boltzmann
Desolvation of polar groups,
Coulomb screening flexibility, entropy terms sampling/Sum over energy wells
Preorganization/Strain
Entropy losses on binding
(rotational, translational, conformational) we count on cancellation of errors within series, or other corrections, which leads to scattered data.
challenge #4
we are not short of idea generators!
easy to construct vast virtual libraries we need more accurate activity prediction to allow filtering and selection
knowledge management
data access tools
learning culture
web2 technologies
build project teams around Sharepoint/OneNote implement a RSS strategy around Newsgator create a literature knowledge sharing culture use Wiki type technology to share knowledge
Pfizerpedia
thanks to
BSA
David Price
Simon Bailey
Julian Blagg
Nigel Greene
CNS MPO
Patrick Verhoest
Travis Wager
Anabella Villalobos
Spiros Liras activity prediction
Marcel de Groot
Martin Edwards
Alex Alex
Jeff Howe
Ben Burke
VLS
Giai Paolini
Willem Van Hoorn
Enoch Huang
Jeff Howe web 2
Jerry Lanfear