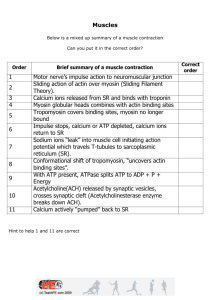
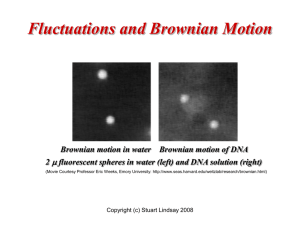
Chapter 10 Nanobiology
advertisement

Nanobiology • • • • Enzymes as nanomachines Molecular motors Fluctuations in gene expression Fluctuations in gene splicing Copyright Stuart Lindsay 2009 The Cell Machine • The genetic material (DNA) is contained inside the nucleus. • Genetic information is transported to the cytoplasm as an RNA copy. • ribosomes translate the RNA code to proteins • the entire cell is packaged in a lipid bilayer membrane. Copyright Stuart Lindsay 2009 Proteins: the nanomachines (A) AP1: Transcription factor binding DNA to regulate the expression of genes (DNA double helix in green) (B)Protein kinase: a dimer that phosphorylates a target (C) Actin, a filamentous protein acting as a structural scaffold. Copyright Lindsay 2009 The 20Stuart amino acid building-blocks Genes code for amino acids The sequence of residues is specified by the sequence of basis in RNA. Each residue is codified by a sequence of 3 bases (codon). The genetic code: Note the stop codons and degeneracy. Note that 42=16 (<20), 43=64 (>20), so a three bases code is necessary to specify the 20 different amino acids. The cell machinery out to the cytoplasm trRNA Into the nucleus Copyright Stuart Lindsay 2009 Mechanical properties of proteins Stiffness Young’s modulus: ratio of stress to strain for a given material. Young’s modulus Stress = Force per unit area F E A Strain = Fractional change in dimension In terms of a Hookean spring (A=l2): F El Spring constant: k = El Ex. Tubulin (transport in the cell) MW=50KDa, E=2GPa, r=2.4nm, k = 10 N·m-1 Ex. Elastin (muscle and ligaments) MW=75KDa, E=0.002GPa, r=2.75nm, k = 0.01 N·m-1 Density Typical density of a well-packed protein is ρ ≈1.4·103 kg·m-3. 3MW r 0.073 MW nm 4 822 3 A (typical) 100kDa protein has a radius of ~3nm. Viscosity Typical viscosity for a globular protein (MW=100kD; r=3nm) is η ≈1·10-3 Pa·s. Drag coefficient in water: 6 r 56 1012 Nsm 1 Diffusion k BT D 6a Typical diffusion coefficient is D ≈7.3·10-11 m2·s-1. Protein motions Long range, collective fluctuations: 6a Damped elastic fluctuations: from ps (tubulin) to ns (elastin) Diffusive fluctuations: tens of ns 2t D Actual catalyzed ET rate is kHz, so only ca. 1 in 106 longrange fluctuations drive the system to the transition state. Copyright Stuart Lindsay 2009 Voltage gated channel Living cells exchange materials by means of channels proteins, chemically selective and under the control of signaling mechanisms. The switch from open to closed is driven by the potential gradient across the cell membrane. AFM images of a monolayer of voltage-gated Porin OmpF on a graphite surface in an electrolyte solution. Energy for Molecular Motors and ATP-dependent enzymes ATP Hydrolysis K eq [ ADP ][ Pi ] 4.9 x10 5 [ ATP] Copyright Stuart Lindsay 2009 Thermal ratchet driven by ATP hydrolysis Reactants = protein motor + H2O Reactants + ATP Products + ADP Products = protein motor one step forward Ea k exp 6a kT Copyright Stuart Lindsay 2009 Molecular motors in muscle cells Motor function in muscle cells is carried out by an actin-myosin complex. The active components of muscle tissues are the sarcomers, thick filaments to which are attached many myosin molecules. • Sarcomeres appear as ca. 100,000 bands in cardiac muscle (TEM image) Sarcomers, thick filaments, are interdigitated with thin filaments, composed of bundles of the actin protein. Crossbridge model Myosin molecules consist of a long stalk that is permanently attached to the tick filament and a pair of head units that transiently contact the actin filaments. The myosin moves along the interleaved actin filaments to draw the crossbridge together, resulting in muscle contraction. Muscle tissues can contract by more than 20% in length on a period of tens of milliseconds. Myosin-actin motor motion Myosin molecule Crystal structure of head unit. Note two “feet”. Arrows point to heads (AAAS) Myosin motor ATP binding, hydrolysis and head motion Actin Filament Copyright Stuart Lindsay 2009 “Walking” action: step is 5 nm (amplified by lever arm to 36 nm). Force is 1.5 pN. A Rotary Motor – ATP Synthase A reversible motor: • Proton gradient drives F1 rotation accompanied by ATP synthesis from ADP. • High ATP concentration drives rotation in opposite direction with ATP hydrolysis which pumps protons. Copyright Stuart Lindsay 2009 clockwise rotation counter-clockwise rotation Copyright Stuart Lindsay 2009 Watching ATP synthase at work F1 unit tethered with dyeloaded actin attached to F0 Movement of the gold bead was detected by laser optical imaging. The motor takes three 120° steps to complete one rotation, hydrolizing one ATP molecule per step. (F1Prop4C.gif) http://www.k2.phys.waseda.ac.jp/Researc.html Copyright Stuart Lindsay 2009 (Courtesy of Professor Kazuhiko Kinosita, Waseda University.) Helix repeat: 3.4 nm Copyright Stuart Lindsay 2008 DNA Nanotechnology About 8 bases must be paired for a double helix to be stable at room Temperature. Copyright Stuart Lindsay 2008 A DNA-based four-way crossover structures producing a rigid planar tile. The distance between adjacent tile is 20nm. The structure, imaged by AFM, is produced by spontaneous selfassembly of the individual crosses. Copyright Stuart Lindsay 2008 DNA Origami A long template strand is annealed with a numebr of short strands that either form cross-links at fixed points (loops) or fill regions to form double helices. Biomimetic nanostructures Mineralization These structures consist of mineral layers held together with proteins that acts as surface-specific ‘glues’ (SEM images). Abalone shell Copyright Stuart Lindsay 2009 Diatom Peptide glues for specific surfaces • Make a random peptide library on the surface of a phage by inserting random DNA into phage genome (Phage display) • Select those phage that stick and grow them up. • Repeat cycle for highly specific interaction • Sequence “successful” phage genome to decode peptide sequence Filamentous bacteriophage sticking to an InP (100) surface. They express a surface protein that sticks to just this particular surface. Whaley, S.R. et al. Nature,2000, 405: 665-668. Mimicking Bio-nano-optics Lenses modeled mimicking brittlestar Brittlestar nanolenses Lee and Szema, Science 2005 but flexible so the refractive index can be adjusted by pumping different fluid.