Powerpoint show
advertisement
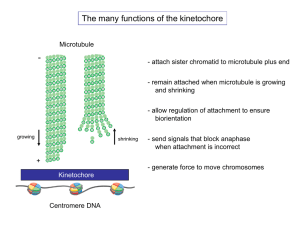
UV Mutagenesis in Yeast Geneticists need variation to study the function of gene products. We create variation in the laboratory by mutagenesis Features of a good model organism •Ease of Genetic Manipulation •Conserved cellular processes •Cell biology •Biochemistry 4 proteins 2 3 5 1 DNA 6 Important lab properties • Genetics: introduce mutations (UV, chemical, Xray) and design screens to identify mutations in process you are interested in • Isolate the products of meiosis • Recover mutations: stable haploid and diploid lifecycles. • Easy molecular manipulations-clonability (few introns), high rate of homologous recombination Fig. 7.2 Mutations are classified by what they do to DNA Fig. 7.6 Fig. 7.12b1 Fig. 7.12b2 By choosing the correct mutagens, we can control the type of mutations we make. Because most of the chemicals are nasty, you will be using UV light to generate mutations Formation of Thymidine Dimer sugar sugar sugar sugar Fig. 7.7 Photoreactivation requires photolyase enzyme Not present in humans Mutagenesis of yeast Haploid Irradiate with UV. Calculate survival curve Select optimal dose for isolation of mutations. Select on appropriate selective media: Replica plating to identify nutrient deficiencies. Survival curve In yeast, 50-90% killing is used in mutagenesis experiments Mutation curve DNA damage is more lethal to haploids Haploids Diploid cells Multicellular diploids Identifying mutants in adenine biosynthesis What can you do with yeast if you are not a geneticist? • Take advantage of the genes identified in genetic screens • Cell biology PCR can generate fusions or point mutations Fusions to GFP allow live cell analysis of protein function Aequoria victoria Green Fluorescent Protein Visualization of chromosome and microtubule dynamics to elucidate mechanisms of nuclear movement and chromosome segregation • lacI-GFP binding to tandem repeat lacO sequences integrated at specific loci. • Dynamics of GFP-fusions to tubulin, spindle pole and mt-based motor proteins. • Specific genetic perturbations to establish cause and effect Yeast Mitotic Spindle Structure EM Tub-GFP Kubai, 1978 Tomographic reconstruction 16 kinetochore microtubules and 4 interpolar microtubules emanate from each spindle pole: 40 mts/1.5 m spindle Architecture of the kinetochore Maiato et al. (2004) CENTROMERE KINETOCHORE MT PLUS ENDS FIBROUS CORONA OUTER PLATE CENTROMERIC HETEROCHROMATIN McEwen et al. (1998) CENP-B MCAK INCENP Aurora B Survivin Borealin/Dasra B ICIS chromatid pairing, structural support, & MT attachment error correction INNER PLATE INTERZONE CENP-A CENP-C CENP-G CENP-H CENP-I/hMis6 CeKNL-1 hMis12 3F3/2 antigens tension receptors & checkpoint signalling Ndc80/Hec1 Nuf2 Spc24 Spc25 BUB1 BUBR1 BUB3 MPS1 MAD1 MAD2 Cdc20 CENP-E KinI,Kip3? kinetochore assembly & size determination MT attachment, regulation of MT dynamics, & checkpoint signalling INNER KINETOCHORE OUTER KINETOCHORE Kinetochores are clustered at the ends the cohesin cylinder Ndc80RFP Spc29RFP Smc3GFP DIC DIC Smc3GFP Ndc80RFP overlay overlay Interstrand cohesin Inflection point Kinetochore sleeve Intrastrand cohesin C-loop Proposed Path of Centromere DNA in a Eukaryotic Kinetochore: C-loop How do we dissect the function of the kinetochore? Pericentric looping requires NDC10 and not MT binding Patterns observed at 37oC and 25oC in temperature sensitive mutants Disruption of Centromeric Attachment via Ndc10 Effects of Centromeric Attachment: ndc10-1 at Restrictive Temperature Centromere Microtubule Disruption of Microtubule Attachment via Nuf2 Effects of Microtubule Attachment: nuf2-60 at Restrictive Temperature Centromere Microtubule Permissive Temperature Restrictive Temperature Effects of Microtubule Attachment on the Inner and Outer Kinetochore Using genetics and cell biology we have dissected functional elements of the kinetochore Inner kinetochore complex CBF3 bends centromere DNA . Clustering of 16 kinetochores requires both inner (COMA, MIND) and outer kinetochore (Ndc80) complexes. Outer kinetochore complexes essential for plus end microtubule interactions. Anderson et al. MCB 2009