- ChemAxon
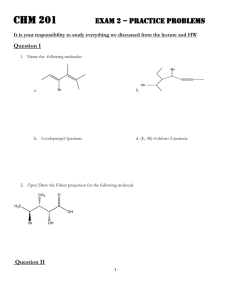
advertisement

Java Solutions for Cheminformatics
Conformer generation
June 2006
The “modeling” team at ELTE
(Eötvös Loránd University)
Ödön Farkas
–
–
–
–
General leadership
Geometry optimization
Fragment fuse
Search involving geometry constraints, etc.
Imre Jákli
– Molecular dynamics (MD)
– Database connection
Adrián Kalászi
– Molecular mechanics
– Drug design tools (3D pharmacophore model)
– Conformer search via MD
Gábor Imre
–
–
–
–
3D builder scheduling
Fragment-atom fuse (v2)
Minkowski-based build
Debug tools
Students: Krisztina Szölgyén, László Antall
May 2006
Conformer generation / basic concepts
• Conformers are locally stable structures of a molecule.
– Conformers are often called “rotamers”, however rings may
also have different conformers which are not rotamers.
• Intermediate structures, corresponding to molecular
motion, are conformations and should not be
considered as conformers.
• The lowest energy conformer can only be found
certainly if all conformers are known.
• The distribution of conformers can be approximated
using the calculated conformational energy.
May 2006
Goal of conformer generation
• Generating valid 3D molecular structures
• Finding multiple structures for flexible molecules
May 2006
History of conformer generation in Marvin
• First approach based on a generalized Minkowski
metric
G. Imre, G. Veress, A. Volford, Ö. Farkas “Molecules from the Minkowski
space: an approach to building 3D molecular structures” J. Mol. Struct.
(Theochem) 666-667, 51 (2003)
• Due to problems with chirality and slow computational
time we introduced an atom-by-atom fuse method
G. Imre, Ö. Farkas “3D Structure Prediction and Conformational Analysis”
7th ICCS, June 5 - 9, 2005 Noordwijkerhout, The Netherlands
• Scheduling is important
• Faster and reliable process
• Frequent use of geometry optimization may slow down the
process
• Current version is based on fusing fragments
May 2006
Key algorithms
used or developed for conformer generation
Quaternion fit (JQuatFit)
• Based on the work of Hamilton
• http://en.wikipedia.org/wiki/Quaternion
• Can fit two molecular structures via non-iterative, linear
scaling, extremely fast method.
• Used for fitting common atoms for fusing fragments
Substructure3DSearch
• Based on the substructure search implemented by ChemAxon
• Simplified for fast exact match (using graph invariant)
• Extended with
• geometry matching (using quatfit) to separate conformers
• high/low priority matching for selecting suitable fuse positions
• geometry constrained topological matching for fragment re-use
• Can quickly distinguish conformers with optional diversity limit
May 2006
Conformer tools in the GUI
MSketch/MView
Draw a molecule
May 2006
Conformer tools in the GUI
MSketch/MView
Draw a molecule
Adjust Clean/3D mode
• Fast build: old algorithm,
no Hydrogens
• Fine build: new algorithm,
automatically adds Hydrogens
• Build or optimize: build only
for non 3D structures
• Optimize: just optimize
Press Ctrl-3 to process
May 2006
Conformer tools in the GUI
MSketch/MView
Pressing F7 changes for
3D rotation mode to
change the viewpoint
Previously Ctrl-F
generated conformers,
now it only displays
if they are available
The new Conformer
plugin is advised for
conformer generation
May 2006
Conformer tools in the GUI
MSketch/MView calculator plugins
The conformer plugin
allows easy access to
the most important
options:
• Output as molecule array or
storage in single molecule
• Variable optimization criteria
• Multiple or single conformer
• Maximum conformer count
• Time limit for the process
• “Hyperfine” mode for thorough
checking of conformers
• H-bond visualization
• Access to old algorithm
May 2006
Conformer tools in the GUI
MSketch/MView calculator plugins
The conformer plugin
allows easy access to
the most important
options:
• Output as molecule array or
storage in single molecule
• Variable optimization criteria
• Multiple or single conformer
• Maximum conformer count
• Time limit for the process
• “Hyperfine” mode for thorough
checking of conformers
• H-bond visualization
• Access to old algorithm
May 2006
Conformer tools in the GUI
MSketch/MView calculator plugins
The conformers can
also be stored as a
property of the molecule
(available in mrv, sdf)
• Single molecule appears
as a result and “Ctrl-F”
displays the stored the
individual conformers
• The desired conformer
to display can be selected
• The selected conformer
should be confirmed.
May 2006
Conformer tools in the GUI
MSketch/MView calculator plugins
The stored conformers
then will appear when
“Ctrl-F” is pressed.
May 2006
Molecular dynamics in the GUI
MSketch/MView calculator plugins
The stored conformers
then will appear when
“Ctrl-F” is pressed.
The flexibility of the
molecule can be studied
via molecular dynamics.
May 2006
Molecular dynamics in the GUI
MSketch/MView calculator plugins
May 2006
Command line conformer tools (cxcalc)
conformers & leconformers
Usage:
cxcalc [general options] [input files/strings] conformers
[conformers options] [input files/strings]
conformers options:
-h, --help
-f, --format
-m, --maxconformers
-s, --saveconfdesc
-e, --hyperfine
-o, --oldalg
-y, --prehydrogenize
-l, --timelimit
-O, --optimization
this help message
<output format> should be a 3D format (default: sdf)
<maximum number of conformers to be generated>
(default: 100)
[true|false] if true a single conformer is saved
with a property containing conformer information
(default: false)
[true|false] if true hyperfine option is set
(default: false)
[true|false] if true old (before Marvin 4.1) algorithm
is used for calculation (default: false)
[true|false] if true prehydrogenize is done before
calculation, if false calculation is done without
hydrogens (available only with old algorithm,
default: false)
<timelimit for calculation in sec> (default: 900)
[0|1|2|3] conformer generation optimiztaion limit
(default: 1)
# cxcalc conformers -m 250 -s true test.sdf
May 2006
Command line molecular dynamics tools (cxcalc)
moldyn
Usage:
cxcalc [general options] [input files/strings] moldyn
[moldyn options] [input files/strings]
moldyn options:
-h, --help
-x, --forcefield
-i,
-n,
-m,
-T,
-j,
this help message
[dreiding] forcefield used for calculation
(default: dreiding)
--integrator
[positionverlet|velocityverlet|leapfrog]
integrator type used for calculation
(default: velocityverlet)
--stepno
<number of simulation steps> (default: 1000)
--steptime
<time between steps in femtoseconds>
(default: 0.1)
--temperature
<temperature in Kelvin> (default: 300 K)
--trajectorytype [mol|sdf] type of output
mol: series of mol frames
sdf: series of sdf frames
(default: sdf)
Example:
cxcalc moldyn test.mol
May 2006
Conformer tools API
// read input molecule
MolImporter mi = new MolImporter("test.mol");
Molecule mol = mi.read();
mi.close();
// create plugin
ConformerPlugin plugin = new ConformerPlugin();
// set target molecule
plugin.setInputMolecule(mol);
// set parameters for calculation
plugin.setMaxNumberOfConformers(400);
plugin.setTimelimit(900);
// run the calculation
plugin.run();
// get results
Molecule[] conformers = plugin.getConformers();
int conformerCount = plugin.getConformerCount();
Molecule m;
for (int i = 0; i < conformerCount; ++i) {
m = conformers[i];
// same as m = plugin.getConformer(i);
// do something with the conformer ...
}
// do something with the results ...
May 2006
3D structure generation capabilities
Comparison
Corina
Much faster…
May 2006
Marvin
15.2 s
3D structure generation capabilities
Comparison
Corina
Much faster…
May 2006
Marvin
5.9 s
3D structure generation capabilities
Comparison
Corina
Much faster…
May 2006
Marvin
5.1 s
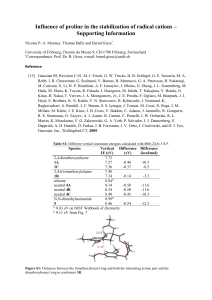
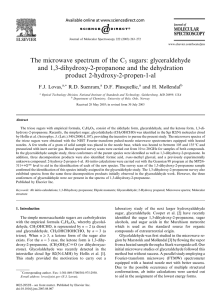
Result statistics
NCI 250K database (August, 2000)
•1st round
• Current method with 120 sec. time limit
• Conversion rate: 99.92% (failed 193 of 250251)
• Avarage time is 0.65 sec/molecule
•2nd round
• Old method on the 193 previously failing structures
• Overall conversion rate: 99.994% (failed 13)
May 2006
Under development
what to expect in the near future
•100% conversion rate for valid,
medium size structures
•Optional conformer diversity limit
•Server version
• Carrying built up fragments for
consequent processes
• Store and use fragment database
•Further speedup
•MMFF94 force field
May 2006
Acknowledgements
May 2006