Species co-occurrences and null models
advertisement

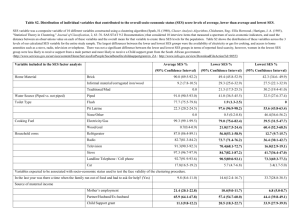
Advanced analytical approaches in ecological data analysis The world comes in fragments Early plant succession in the Saxon (Germany) post brown coal mining area Chicken Creak 2005 2010 Succession starts with colonising species from a regional species pool and from the initial seed bank Species co-occurrences How do patterns of species co-occurrences change in time? 2005 Agrostis_capillaris Agrostis_stolonifer Cirsium_arvense A1-2 0 0.1 0.1 A3-2 0 0 0.5 Basic questions 1. Which species colonise initially? 2. Is initial colonisation directional? 3. Is colonisation predictable? A4-2 0.1 0 0 A4-3 0 0 0 B1-2 0 0 0.1 B2-1 0 0.1 0 B2-4 0 0.1 0 Starting hypothesis 1. Island biogeography predicts initial species occurrences to be random. 2. Compretition theory predicts equilibrium species occurrences to be not random but driven by interspecific competition. Early plant succession 2011 Achillea_pannonica Agrostis_capillaris Agrostis_stolonifer Agrostis_vinealis Ajuga_genevensis Apera_spica-venti Arenaria_serpyllifo A1-1 0 0 0 0 0 0 0 A1-2 0.5 0.5 0 0 0 0.4 0 A1-3 0 0 0.5 0 1.5 0 0 A1-4 0 0.5 0 2.0 0 0 0 A2-1 3.0 0.5 0 0 0 0.7 0.5 A2-2 0.1 0.5 0 0 0 0 0.5 A2-3 0 0.5 0 0 0 0 0.1 How to test for (non-) randomness? Habitat filtering Species Joined absences 1 A 0 B 0 C 0 D 1 E 0 𝐶𝑙𝑢𝑚𝑝𝑖𝑛𝑔 𝑠𝑐𝑜𝑟𝑒 = 2 1 0 0 0 1 3 0 1 0 0 0 Sites 4 5 0 0 0 1 0 1 0 0 0 0 Reciprocal segregation Checkerboard 𝐶 − 𝑠𝑐𝑜𝑟𝑒 = 7 1 1 0 1 1 8 1 1 0 1 1 Clustered co-occurrence Joined co-occurrence Segregation Competition Reciprocal habitat requirements 6 0 0 0 1 0 4 𝑐𝑙𝑢𝑠𝑡𝑒𝑟 𝑚𝑛(𝑚 − 1)(𝑛 − 1) Common ecological requirements Habitat engineering by the earlier colonizer 4 𝑐ℎ𝑒𝑐𝑘𝑒𝑟𝑏𝑜𝑎𝑟𝑑𝑠 𝑚𝑛(𝑚 − 1)(𝑛 − 1) Habitat filtering Niche conservatism Facilitation, mutualism Niche conservatism is the tendency of closed related species to have similar ecological requirements and life history raits Species Spatial species turnover (b-diversity) A B C D E 1 0 0 0 1 0 2 1 0 0 0 1 3 0 1 0 0 0 Sites 4 5 0 0 0 1 1 1 0 0 0 0 6 0 0 0 1 0 7 1 1 0 1 1 8 1 1 0 1 1 EV -0.5 0.26 2.16 -0.6 -0.5 Species EV -0.7 -0.6 0.3 2.4 1.4 -0.7 -0.4 -0.4 Sort according to the dominant eigenvector of correspondence analysis C1 B2 A3 E4 D5 41 1 0 0 0 0 52 1 1 0 0 0 33 0 1 0 0 0 Sites 74 85 0 0 1 1 1 1 1 1 1 1 26 0 0 1 1 0 17 0 0 0 0 1 86 0 0 0 0 1 2.4 1.4 0.3 -0.4 -0.4 -0.6 -0.7 -0.7 2.16 0.26 -0.5 -0.5 -0.6 Sorting a presenceabsence matrix according to the dominant eigenvectors of corresondence analysis (seriation) maximizes the number of occurrences along the left to right diagonal. Metric for spatial species turnover Squared correlation R2 of row and column ranks of species occurrrences. R 1 1 2 2 2 2 3 3 3 4 4 4 5 5 5 5 C 1 2 2 3 4 5 4 5 6 4 5 6 4 5 7 7 Pearson 0.75 R2 0.56 Species A nested subset pattern A B C D E 1 1 1 1 1 1 2 1 1 1 1 0 Sites 3 4 5 1 1 1 1 1 1 1 0 0 0 0 0 0 0 0 6 1 0 0 0 0 7 1 0 0 0 0 8 1 0 0 0 0 Nestedness describes a situation where a species poorer site is a true subset of the next species richer site. Sum 8 5 3 2 1 A presence – absence matrix ordered according to total species richness (marginal totals, degree distributions) Unexpected absence Unexpected presence Species Sum 5 4 3 2 2 1 1 1 A B C D E 1 1 1 1 1 1 2 1 0 1 1 0 Sites 3 4 5 1 1 1 1 1 1 1 0 0 0 0 0 0 0 0 6 1 0 0 1 0 7 1 0 0 0 0 8 1 0 0 0 0 Sum 5 3 3 2 2 2 1 1 Sum 8 4 3 3 1 The measurement of nestedness The distance concept of nestedness. A 1 1 1 1 1 0 1 1 1 1 1 1 0 1 0 0 0 0 0 0 C 1 1 1 1 1 0 1 0 0 0 0 1 1 0 1 0 0 1 1 0 1 1 1 0 1 0 1 1 0 0 1 0 1 0 0 1 1 0 0 0 M O P D F H K E B J N L G 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 D 1 1 1 1 1 1 1 0 1 1 1 1 1 1 0 1 0 1 1 0 1 1 20;P 1 1 0 0 1 1 0 1 1 0 1 1 1 0 1 1 1 1 1 0 0 0 0 0 d 1 0 1 1 1 1 1 1 0 1 1 0 0 1 0 1 0 0 1 1 0 d 1 13;J 0 1 0 0 1 0 1 1 1 1 0 1 0 1 0 0 0 1 1 X;Y 0 0 1 1 0 1 0 0 0 1 0 1 1 0 0 0 1 0 0 0 1 0 1 0 1 1 0 0 1 0 1 0 0 0 0 0 0 0 1 1 0 1 0 0 0 0 0 0 0 1 0 0 1 1 0 0 0 0 1 0 0 0 0 0 0 0 1 0 0 0 0 1 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 D0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 Sum 12 11 10 10 10 10 9 9 100 1 𝑇= 0.04145 𝑚𝑛 9 8 7 𝑚 𝑛 1 1 6 6 𝑑𝑖𝑗 𝐷𝑖𝑗 6 5 2 Sum 16 16 12 11 10 9 9 9 7 6 6 6 5 3 2 1 1 1 1 1 Sort the matrix rows and columns according to some gradient. Define an isocline that divides the matrix into a perfectly filled and an empty part. The normalized squared sum of relative distances of unexpected absences and unexpected presences is now a metric of nestedness the nestedness temperature. 4 Z-score I 1 3 7 15 20 4 9 13 2 11 17 18 5 8 16 6 10 12 14 19 8 6 4 2 0 -2 -4 -6 -8 0 50 Matrix size 100 Nestedness based on Overlap and Decreasing Fill (NODF) r1 c2 1 0 c3 1 c4 c5 1 Nestedness among columns c1 1 r2 1 1 1 0 0 r3 0 1 1 1 0 r4 1 1 0 0 0 r5 1 1 0 0 0 Nestedness among rows c1 c2 c1 c3 c1 c4 c1 c5 c2 c3 1 0 1 1 1 1 1 1 0 1 1 1 1 1 1 0 1 0 1 1 0 1 0 1 0 1 0 0 1 1 1 1 1 0 1 0 1 0 1 0 1 1 1 0 1 0 1 0 1 0 Npaired=0 Npaired=67 Npaired=50 Npaired=100 Npaired=67 c2 c4 c2 c5 c3 c4 c3 c5 c4 c5 0 1 0 1 1 1 1 1 1 1 1 0 1 0 1 0 1 0 0 0 1 1 1 0 1 1 1 0 1 0 1 0 1 0 0 0 0 0 0 0 1 0 1 0 0 0 0 0 0 0 Npaired=50 r1 1 0 1 1 1 r2 1 1 1 0 0 r1 1 0 1 1 1 r3 0 1 1 1 0 r1 1 0 1 1 1 r4 1 1 0 0 0 r1 1 0 1 1 1 r5 1 1 0 0 r2 0 1 1 r3 1 1 0 r2 0 1 1 1 0 r4 1 1 0 0 0 r2 1 1 0 0 0 r5 1 1 0 0 0 r3 1 1 1 0 0 r4 0 1 1 1 0 r3 1 1 1 0 0 0 r5 1 1 0 0 0 1 0 r4 1 1 1 0 0 0 0 r5 1 1 0 0 0 Npaired=67 Npaired=67 Npaired=50 Npaired=50 Npaired=50 Npaired=0 Npaired=100 Npaired=100 Npaired=100 Npaired=50 Npaired=0 Ncolumns = 63.4 Npaired=0 Nrows = 53.4 Npaired=100 NODF = 58.4 Npaired=100 2 𝑁𝑝𝑎𝑖𝑟𝑒𝑑 𝑁𝑂𝐷𝐹 = 𝑚 𝑚 − 1 + 𝑛(𝑛 − 1) NODF is a gap based metric and more conservative than temperature. Back to the Huehnerwasser How does species co-occurrence change during early succession? 0.008 0.006 0.5 0.4 0.3 0.004 0.2 0.002 0.1 0 2005 2006 2007 2008 2009 2010 2011 1 0.8 0.6 The degree of nestedness is at an average level (neithert nested nor antinested) 0 2005 2006 2007 2008 2009 2010 2011 Study year Study year R2 C-score 0.01 0.6 The number of reciprocal species cooccurrences increases in time NODF 0.012 The degree of species spatial turnover decreases in time 0.4 0.2 0 2005 2006 2007 2008 2009 2010 2011 Study year But are raw scores reliable? What do we expect if colonisation were a simple random process? Statistical inference using null models Species Species What is random in ecology? 1 01 0 0 01 0 2 01 01 0 0 0 3 0 0 0 0 0 Sites 4 5 0 0 0 1 0 0 0 0 0 0 0 Sum 5 4 3 2 6 1 0 0 0 0 7 0 0 1 0 0 8 1 0 0 0 0 1 1 1 A B C D E 2 1 1 1 1 1 1 2 1 1 1 1 0 3 1 1 0 1 0 Sites 4 5 1 0 1 1 0 1 0 0 0 0 Sum 5 4 3 2 A B C D E 2 6 10 0 0 10 0 7 0 0 0 0 0 8 0 0 0 0 0 1 1 1 Sum 8 5 3 2 1 The proportionalproportional null model Fill the matrix at random but proportional to observed marginal (row/column) totals until the observed total number of occurrences is reached The fixed - fixed null model Sum 6 5 3 3 1 Fill the matrix at random until for each row and each column the observed total number of occurrences is reached Take a checkerboard pair and swap. Do this 100000 times to randomize the matrix. Statistical inference using null models Frequency 0.30 120 400 randomized matrices 0.25 100 Observed score 0.20 80 0.15 60 - SES SES 0.10 40 0.05 20 0.00 0 0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 1 Score class For each randomized matrix we calculate the respective metric (C-score, NODF, R2). 𝑆𝐸𝑆 = (𝑂𝑏𝑠 − 𝐸𝑥𝑝) 𝑆𝑡𝐷𝑒𝑣 Standardized effect sizes (SES) are Ztransformed scores and can be linked to a normal distribution. The Fisherian significance levels P(-1 < X <+1) = 68% P(-1.65 < X < +1.65) = 90% P(-1.96 < X < +1.96) = 95% P(-2.58 < X < +2.58) = 99% P(-3.29 < X < +3.29) = 99.9% Year Score Average null distribution score Standard deviation Z-score (SES) FF null model P(Z) 0.00009 0.00002 0.00001 0.00001 0.00001 0.00001 0.00001 0.193 1.106 3.036 4.995 4.454 7.273 10.273 0.847184 0.26857 0.002319 6E-07 7.9E-06 0 0 0.00575 0.00116 0.00171 0.00274 0.00122 0.00081 0.00089 -0.12 -1.095 -0.138 -1.358 0.083 -0.498 0.053 0.904851 0.27362 0.890053 0.174224 0.933961 0.618628 0.95803 0.02176 0.04284 0.02361 0.011 0.00509 0.00507 0.00497 -0.523 -0.168 1.238 6.025 6.705 5.463 2.488 0.601039 0.866252 0.215409 0 0 1E-07 0.012634 C-score 2005 2006 2007 2008 2009 2010 2011 0.01087 0.00106 0.00132 0.00355 0.00492 0.00756 0.00813 0.01085 0.00104 0.0013 0.00349 0.00487 0.00747 0.008 NODF 2005 2006 2007 2008 2009 2010 2011 0.05931 0.48707 0.53815 0.46488 0.49842 0.52119 0.49632 0.05999 0.48834 0.53838 0.4686 0.49832 0.52159 0.49627 R2 2005 2006 2007 2008 2009 2010 2011 0.91608 0.27526 0.20723 0.12074 0.06106 0.04678 0.02964 0.92746 0.28248 0.17799 0.05448 0.02695 0.01907 0.01728 8 6 4 2 0 2005 2006 2007 2008 2009 2010 2011 Study year SES R2 SES C-score 10 SES NODF 12 SES scores (FF mull model) The degree of reciprocal 0.2 species segregation 0 constantly increases -0.2 -0.4 during early succession -0.6 Local plant communities -0.8 are not significantly -1 nested during succession -1.2 -1.4 -1.6 2005 2006 2007 2008 2009 2010 2011 Study year 8 7 Species spatial turnover 6 peaks at intermediate 5 stages of succession 4 3 2 1 In ecological research 0 raw metrics are most -1 often meaningless!! 2005 2006 2007 2008 2009 2010 2011 Study year 0.2 0 -0.2 -0.4 -0.6 -0.8 -1 -1.2 FF -1.4 -1.6 2005 2006 2007 2008 2009 2010 2011 3 2 1 0 -1 -2 2005 2006 2007 2008 2009 2010 2011 Study year 12 1 10 0.5 SES C-score SES C-score Study year 8 6 4 2 FF 0 2005 2006 2007 2008 2009 2010 2011 Study year PP 4 SES NODF SES NODF Different null assumptions give different answers 0 -0.5 -1 -1.5 PP -2 2005 2006 2007 2008 2009 2010 2011 Study year Study year The FF null model assumes that sites are filled with species and that each species occupies the maximal number of sites. 5 4 3 SES R2 SES R2 8 7 6 5 4 3 2 1 0 FF -1 2005 2006 2007 2008 2009 2010 2011 2 1 0 -1 PP -2 2005 2006 2007 2008 2009 2010 2011 Study year The PP null model assumes that sites might differ dynamically in species richness and that each species might occupy a variable number of sites. The PP null model points often to a random pattern in species occupancy. Idiosyncratic species are those that deviate in their patterns of occurrences Total NODF ExpNODF SpResult Z-SpResu DeltaSpC Occ Cirsium_arvense 11 56.618 63.431 6.087 0.864 -2.229 Chenopodium_album_agg. 7 69.118 69.402 5.975 1.049 -1.146 Species 2005 6 6 6 75 91.176 66.912 74.615 79.348 80.944 5.409 5.365 6.245 -0.529 -0.607 3.091 -1.452 -1.382 -0.536 3 86.029 86.852 5.919 1.163 -0.05 3 94.118 85.908 5.311 -1.004 -0.841 Arenaria_serpyllifolia_agg. Festuca_rubra_agg. Daucus_carota Agrostis_capillaris Carex_ericetorum Bromus_tectorum Robinia_pseudoacacia Lupinus_luteus Poa_palustris Rubus_fruticosus_agg. 2 2 2 2 2A 2 B 1 C 1 D 1 E 0 92.647 85.294 92.647 92.647 1 2 98.529 1 1 92.647 1 1 92.647 1 1 92.647 1 1 92.647 1 0 100 90.293 90.795 92.899 Sites 3 91.203 4 5 6 1 91.166 1 1 1 1 91.345 1 1 0 96.805 1 1 0 0 0 96.949 0 0 1 94.787 0 0 0 0 100 5.859 5.97 5.505 75.831 8 15.792 1 05.822 0 5.838 0 0 15.789 0 5.816 0 1 5.629 1.341 1.683 -0.527 1.133 1.035 0.806 1.672 1.126 1.269 0 -0.008 0.075 -0.383 -0.092 -0.133 -0.092 0.075 0.075 0.075 0.075 Species Agrostis_stolonifera_agg. Crepis_tectorum Echium_vulgare Rumex_acetosella_var._te nuifoliu Calamagrostis_epigejos Cirsium arvense increases (DeltaC < 0) the degree of nestedness None of the species decreases the nestedness pattern. There are few unexpected occurrences . All species behave similar.