Crtystallography
advertisement

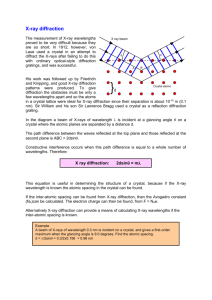
Macromolecular crystallography X-ray Crystal Model building Phasing Diffraction pattern Electron density map Atomic structure model What we can get from the structures of biological macromolecules? 1. Interaction of protein-protein or protein-biomolecule 2. Mechanism of enzyme 3. Mechanisms of biological functions 4. Drug design 5. Bases for molecular dynamics simulations, protein design and engineering The History of X-Ray Crystallography in the Eyes of Nobel Prizes http://www.ibric.org/myboard/print.php?Board=report&id=2311 Structures of biological macromolecules 102,863 structures (2014. 8. 26) are deposited in PDB Among those, 88.7 % are X-ray crystal structures, 10.3 % are solved by NMR and 0.8 % are by EM Crystallization of macromolecule (protein) What really looks like inside of crystal? • • • • • • 3X3 2D grid of protein crystal Black box; unit cell Red box; asymmetric unit Infinite extension of unit cell on XYZ axis Empty space are filled with solvent (water) Average solvent contents of protein crystals ; 50% more or less X-ray diffraction; special case of x-ray scattering X-rays: 0.1 Å < λ < 1000 Å (1 Å = 10-10 m = 100 pm = 0.1 nm) atomic distances (d): ~ 1.5 Å Ex) visible light; 400 - 700 nm Maximum resolution of diffraction data can be determined by y/D (θ); dmin=mλD/ymax X-ray diffraction; crystal vs diffraction pattern Diffraction pattern 2D crystal Reciprocal relationship Fourier transform 2D crystal Reciprocal relationship Structure of molecule in crystal can be determined from diffraction pattern by Fourier transformation Calculating electron density; Fourier transformation Structure factor Electron density h,k,l=reciprocal lattice x,y,z=coordinate of real space Electron density Fourier transform Intensity of diffraction spot Phase Electron density Now Phase problem!!! Patterson function; interatomic relationship within the unit cell • Calculation of x,y,z coordinates of atoms is possible in special position of Patterson space (u,v,w), where u, v, or w has defined value (0, 1, 1/n etc. ; Harker section) • Indeed, it is possible to determine all the positions of atoms when the number of atoms are enough small and distinguishable ( < 100 and high difference in electron density) • But in case of protein, atoms are usually more than 1000 and the differences of electron densities are not distinguishable (C, N, O, S) What if we can have distinguishable atoms in protein crystal? Patterson map of heavy atom derivative • Heavy atoms such as Hg, Au, Ag, Ur Pt can bind to the protein with some degree of specificity without disturbing structure or crystal packing in certain conditions • Amplitude of heavy atom in FH can be obtained from experiment • With the known electron density and coordinate obtained from Patterson function, Phase of heavy atom can be calculated Vector presentation of structure factor FPH = FP + FH Multiple isomorphous replacement (MIR) FPH First derivative FP FH second derivative Multiple anomalous dispersion (MAD) incident photon with relatively low energy • • • incident photon with high enough energy Anomalous scattering maximized at absorption wavelength Synchrotron is needed for tunable wavelength Due to the Se-methionine derivate generation by auxotroph, most of the novel structures solved recently by MAD (SAD) Molecular Replacement (MR) • With using similar structure, calculating phase of unknown structure How can I confirm that I solve the right structure in MR? FT + = Model building and refinement Resolution vs Model Validation of structure • R-factor represents the ideality of the model • R-free represents the correlation of model with experimental data (electron density) • Validation of stereochemical ideality of model • Residues should not be in disallowed region (white) when the resolution of map is better than 3.0 A