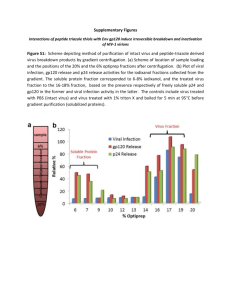
Production and Glycosylation Analysis of Model Proteins
advertisement

Production and Glycosylation Analysis of Model Proteins from a Vaccinia Virus-Mammalian Cell Expression System Nicole A. a a,b,c Bleckwenn , William b Biotechnology Unit, NIDDK, National Institutes of Health, DHHS, Bethesda, MD Abstract hGC-1 Protein (intracellular) Western Blots Fluorescent Microscopy Images + EGFP-6xHis gene Induced 60 42 42 30 22 17 30 22 17 + EGFP-6xHis gene Not Induced Vacuum Mesh Screen Module Anti-FLAG antibody Water Jacket *** ATF™ Secreted gp120-6xHis (mg/L) 40 30 10 0 50 60 70 Wash column with base buffer (50 mM NaH2PO4 300 mM NaCl pH 8.0) Time (hpi) Western blot bands Quantification achieved by scanning blot and comparing to gp120 standard**** with Scion Image software, Scion Corp. ****gp120 standard, HIV-1SF162 gp120 from Chiron Corporation and the DAIDS was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. Deglycosylation Analysis 1 2 3 4 5 Treatment ATF Controller Air Inlet pVOTE.2* Plasmid virus and pVOTE.2 plasmid were kindly supplied by B. Moss and P. Earl, NIAID, NIH (4) and gp120 plasmid pTM-DHgp120H was kindly provided by M. Cho, School of Medicine, Case Western Reserve University (5). Production Parameters for microcarrier perfusion culture Cleavage of N-linked Sugars PNGase F – Cleaves most N-linked oligosaccharides (unless a(1-3) core fucosylated) Cleavage of Common O-linked Sugars O-Glycosidase – Cleaves O-linked unsubstituted Gal-b(1-3)GalNAc-aa-2(3,6,8,9) Neuraminidase – Cleaves non-reducing terminal branched and unbranched sialic acids Cleavage of less common hexasaccharide structures b(1-4)Galactosidase – Cleaves b(1-4)-linked, non-reducing terminal galactose b-N-Acetylglucosaminidase – Cleaves b-linked Nacetylglucosamine Treat to deactivate virus 0.5% NP-40 500 mM NaH2PO4 300 mM NaCl Incubate overnight 4°C Deglycosylation Results kDa 250 148 Std 60 42 30 22 17 6 4 • Cell Growth • 5 g/L Cytodex 3 microcarriers • 1.5x105 HeLa cell/mL initial seeding • DMEM+10% fetal bovine serum • 37°C, 30% dissolved oxygen, pH 7.0 • Growth for 5 days to 1-2x106cell/mL • Infection • 1 hour duration • MOI 5.0 (pfu/cell) • One third working volume • No serum in infection media (DMEM) • 1 mM IPTG added at infection • Production • Return to 1.5 L volume and 10% serum • Reduce temperature to 34°C • Increase dissolved oxygen to 50% • 66 hours post infection gp120-6xHis Purification 48 hpi Reactor Supernatant • Bind protein • Recycle supernatant through column for 4 hours • Wash column • 1X with base buffer • 2X with base buffer + 20 mM imidazole • Elute protein • Four fractions with base buffer + 200 mM imidazole Time (hpi) 18 24 30 40 44 48 52 66 Aliquot five fractions and treat sequentially with enzymes Recombinant virus * vT7lacOI Purification Methods 20 gp120 purified from reactor at 48 hpi in 10 mM Tris pH 7.0 Donor Plasmid ATF™ System*** Clarify by centrifugation 40 Gene System was kindly supplied by Refine Technology, Co., East Hanover, NJ. Load column with 50% slurry Ni-NTA resin (Qiagen) 1 mL resin per 100 mL supernatant cell/mL infected at 0 hpi 30 • Plaque isolation • Amplification • Viral purification • Viral titer Recombinant Plasmid antibody 50 20 Gene Inlet Outlet gp120-6xHis Production 10 Transfect • HeLa cells (attachment dependent) • Microcarrier perfusion culture • 1.5 L working volume reactor HIV-1 gp160 Antiserum (HT3) from DAIDS, NIAID, NIH produced under contract by Repligen was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. 0 HeLa Cells Gas inlet into reactor headspace Feed Pump ** Anti-gp160 Antibody, 1.4x106 Filtrate Pump DO pH Temp. Agit. Level Control Anti-gp160** Infect Based on reporter protein (EGFP) process development 6 4 6 4 Department of Chemical Engineering, UMCP, College Park, MD vT7lacOI* virus gp120 Protein (extracellular) kDa 250 148 60 a Shiloach Virus Construction Defining the Culture System Diaphragm No virus c Center for Biosystems Research, UMBI, College Park, MD • Vaccinia Virus • Orthopoxvirus family Poxviridae • Transcription occurs in cytoplasm of infected cell • Wide host range, includes most mammalian species and humans • HeLa cells • Attachment dependent strain (ATCC CCL-2) • Microcarrier growth for larger cultures • Enhanced Green Fluorescent Protein (EGFP) • Used as reporter protein to develop system parameters • hGC-1 • Olfactomedin-related protein (~64 kDa) • Potential for treatment of prostate and other cancers • Six N-linked glycosylation sites • gp120 • HIV envelope coat protein (~120 kDa) • Almost half the molecular weight attributed to N-linked glycosylations which are required for activity of the protein (3) Expression Verification kDa 250 148 and Joseph Background A vaccinia virus-mammalian cell expression system was developed as an alternative method for recombinant protein production utilizing EGFP as a reporter protein (1). In previous work, EGFP production was evaluated in T-flask culture and in both suspension and microcarrier based bioreactor systems, where general production parameters were defined. In this work, the production capability of the system, as defined with EGFP, was evaluated using two proteins, the HIV gp120 envelope glycoprotein and hGC-1 (2), an olfactomedin-related protein. These proteins contain complex post-translational modifications, required for gp120 activity and possibly for hGC-1, although there is little information on this recently discovered protein. Two recombinant vaccinia virus strains were engineered with the genes for gp120 or hGC-1 and expression of these proteins was achieved in either T-flask culture or both T-flask and bioreactor culture by infection of the cell culture with recombinant virus. The production process, purification protocol and glycosylation pattern of gp120 is described. Bioreactor culture produced secreted gp120 up to 40 mg/L at 66 hours post infection (hpi). EGFP Reporter Protein (intracellular) b,c Bentley , Treatment 1 – No enzymes Treatment 2 – PNGase F Treatment 3 – PNGase F O-Glycosidase Treatment 4 - PNGase F O-Glycosidase a-2(3,6,8,9) Neuraminidase Treatment 5 - PNGase F O-Glycosidase a-2(3,6,8,9) Neuraminidase b(1-4)Galactosidase b-N-Acetylglucosaminidase Coomassie Stained Gel Western Blot kDa 250 148 60 42 30 22 17 6 4 Conclusions • Three different proteins were produced from three constructed viruses (EGFP, hGC-1, and gp120) • Growth, infection and production parameters were defined using reporter protein EGFP • gp120 was produced in a 1.5L bioreactor culture over 8 days with production for 66 hours • Per liter of working volume • 4.7 mg secreted and purified gp120 was recovered at 48 hpi • 4 liters of media used • 200 mL serum used • 370 mL viral stock used (1.9x1010 pfu/mL) • Deglycosylation analysis of gp120 showed significant amount of Nlinked glycosylations of approximately half the mass of the protein • Similar analysis will be performed on bioreactor culture with hGC-1 containing virus References 1. Bleckwenn, N.A., W.E. Bentley, and J. Shiloach, Biotechnology Progress, 2003. 19(1): p. 130-136. 2. Zhang, J.C., et al., Gene, 2002. 283(1-2): p. 83-93. 3. Hu, Y.C., et al., Biotechnology Progress, 2000. 16(5): p. 744-750. 4. Ward, G.A., et al., Proceedings of the National Academy of Sciences of the United States of America, 1995. 92(15): p. 6773-6777. 5. Lee, M.K., M.A. Martin, and M.W. Cho, Aids Research and Human Retroviruses, 2000. 16(8): p. 765-775.