L7_DE
advertisement
Differential Expressions
Classical Methods
Lecture Topic 7
What’s your question?
• What are the genes that are different for the healthy versus diseased
cells?
– Gene discovery, differential expression
• Is a specified group of genes all up-regulated in a specified condition?
– Gene set differential expression
• Can I use the expression profile of cancer patients to predict
chemotherapy outcome?
– Class prediction, classification
• Are there tumour sub-types not previously identified? Do my genes
group into previously undiscovered pathways?
– Class discovery, clustering
• This lecture covers first question - differential expression.
Some terms
• The following terms are used exchangeably:
• Differentially versus not differentially expressed
• Active versus Inactive genes
• Affected versus not affected genes
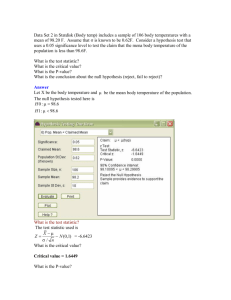
• We will first deal with the simplest case, comparing 2
conditions (say treatment and control).
• We will also mainly be interested in hypothesis testing
(HT) aspect of Inference
Inference
• Lets first differentiate between the different
schools of thought:
• Classical Inference
• Bayesian Inference
• Empirical Bayes’ methods
The MAIN Difference
• The Classical School believes that PARAMETERS are
unknown but FIXED quantities and the data are random
• The Bayesians believe after observing, data can be
considered fixed, whereas our knowledge about the
parameter is truly random.
• This leads to two vastly different APPROACHES when it
comes to hypothesis testing, though both approaches are
inherently interested in the parameter.
Consider the following situation
qg= population mean difference in gene expression from
treatment to control for gene g.
• Both approaches are interested in this. Particularly, both
approaches are interested in seeing if the parameter is 0 or
not, i.e. whether or not the genes are differentially
expressed.
• This corresponds to information about v I
g
{q g 0}
The difference
• Classical Statisticians tend to be drastic about the choice of I. It is
either 0 or 1.
• Bayesians on the other hand tend to describe this value with a
probability, just as if it were another parameter.
• The classical approach to (HT) in micro-array is to assume the
negative hypothesis (null or H0) and try to find the absurdity
probability of the null hypothesis.
• The Bayesian approach assumes that for each gene g, there is the
unobservable function, vg, that defines the genes activation status. The
idea here is to update the knowledge of vg in terms of a posterior
probability. This is the probability that vg=0.
The Classical Procedure
• Here we have some claim or belief or knowledge
or guess about a population parameter which we
want to prove (or disprove) using our data for
evidence.
•
• Hypothesis is always written as a pair.
• Research: what we are trying to prove (Ha)
Null: that nullifies / negates our research.(H0)
The hypotheses
• H0g: gene g is NOT differentially expressed
• H1g: gene g IS differentially expressed.
• We have n pairs of mutually exclusive hypothesis.
• The role of the two hypothesis is NOT symmetric.
Classical Statistics assumes the null by default UNLESS
enough contrary evidence is found.
• Hypothesis is a systematic procedure to summarize
evidence in the data, in order to decide between the pair of
hypothesis.
The Test Statistic
• The test statistic is a summary of the data used to evaluate
the validity of the hypothesis.
• The summary chosen depends upon the question and
circumstances.
• Typically the Test Statistic is chosen, so that when H0 is
true the distribution of the test statistic has a certain known
distribution.
The logic of hypothesis testing
• To actually test the hypothesis, what we try to do is to
disprove or reject the null hypothesis.
• If we can reject the null, by default our Ha (which is our
research) is true.
• How do we do this?
• We take a sample and look at the test-statistic. Then we
try to quantify that, if the null was true, would our
observed statistic be a likely value from the known
distribution of test statistics.
• If our observed value is not a likely value, we reject the
null.
• How likely or unlikely a value is, is determined by the
sampling distribution of that statistic.
Error Rates
• Since we take our decisions about the parameter based on sample
values we are likely to commit some errors.
• Type I error: Rejecting Ho when it is true
• Type II error: Failing to reject Ho when Ha is true.
• In any given situation we want to minimize these errors.
• P(Type I error) = a, Also called size, level of significance.
•
• P(Type II error) = b,
• Power = 1-b, HERE we reject Ho when the claim is true. We want
power to be LARGE
Terms in error control
• In hypothesis we have two main types of error:
• Null hypothesis is wrongly rejected (FALSE POSITIVE)
• Null hypothesis is wrongly accepted (failed to reject)
(FALSE NEGATIVE).
• Trade off is one cannot control both probabilities of both
errors together.
• Size = Probability of a False Positive
• Power= 1 – Probability of a False Negative
Decision Rule
• A decision rule is a method than translates the vector of
observed values into a binary decision or REJECT or FAIL
TO REJECT.
• The decision rule is made such that the error rate of Type I
error is controlled.
• P-value: the absurdity value of the null hypothesis, or the
probability of observing a number more extreme than the
observed value, assuming that the null is true.
Classical Test for differential Expression
concerning two populations
• The two sample t test
–
–
–
–
–
–
Version 1: The Pooled t test
Version 2: The Satterwaite Welch’s t test
Version 3: Bootstrapped t tests
Version 4: Permutation t test
Wilcoxon Rank-Sum test (nonparametric alternative)
Likelihood Ratio Test
The Test Statistic: In general
• Notation: Let xgi1,…,xgik represent the observations
(intensity/ log intensity/ normalized intensity etc) for gene
g, for the ith condition with, i=1,2. Let the means of the
two conditions, the standard deviations and the variance be
given by
2 2
x1g ., x2 g ., s1g , s2g
t-test
• The test statistic is given by:
( x1 g x2 g )
t
1
1
se(
)
k1 k 2
se
(k1 1) s12g (k2 1) s22g
(k1 k2 2)
, pooled t
This follows a t distribution with (k1+k2-2) df
The Satterwhite-Welch’s (SW) Test Statistic
t
( x1g x2 g ) ( 1 2 )
2
s1g
k1
df
2
s 2g
follows a t with
k2
(k1 - 1)(k 2 - 1)
(k 2 1)c 2 (1 c) 2 (k1 1)
where c
2
s1g
/ k1
2
2
s1g / k1 s 2g / k2
The distribution of the Test Statistic
• The pooled t assumes that the two variances are
equal. The SW test does not.
• Both assume underlying normality for the
observations xgi1…xgiki.
Bootstrapping and Permutation t Tests
• Before we do the tests lets talk about the procedures.
• The tests use the principles of Bootstrapping or
Permutation and apply it in the case of differential
expression for two conditions.
Bootstrapping
• The term “bootstrap” came from the legendary Baron
Munchausen, who pulled himself out of a man-hole
literally by grabbing his own bootstraps.
• Efron (1979) introduced the idea into Statistics, the idea is
to “get yourself out of a hole” by re-using the data many
times.
• It is essentially a re-sampling technique that simulates
alternative values of the statistic under the null (in our case
the t statistic), in order to calculate an empirical p-value.
• The idea is to create a large number of values from the
statistic to get an idea of the distribution of the test statistic
UNDER the null.
Bootstrapping contd…
• First I will use a small example for illustrating the
procedure and then I will introduce notation.
• Lets say we have data on gene “g” for condition 1 and
condition 2 as follows:
• Cond1
• Cond2
12
15
15
32
24
26
17
24
Bootstrapping
• Calculate the observed t value for your data
• First you pool the data:
• Then samples of size 4 with replacement from the pooled sample, one
for sample 1 and the other for sample 2. Calculate the t statistic
• Continue this for B samples.
• Find what proportion of observations are BIGGER than your observed
value. This is your “estimated p-value” using bootstrap.
#{| stat || obs |
p value
# samples
Example
s1
12
15
24
17
s2
15
32
24
17
comb
12
15
24
17
15
32
24
17
s1b
15
17
12
24
s2b
17
24
17
24
Histogram of Bootstrapped t values
-1.08
180
160
140
Frequency
120
100
80
60
40
25
20
0
-6.0
-4.5
-3.0
-1.5
boot
0.0
1.5
3.0
Permutation Test
• Similar Idea to Bootstrapping.
• Here instead of resampling WITH replacement we
“shuffle” the data to look at all possible permutations.
• Hence, we pool the data, then sample without replacement
from the pooled data.
• Same Idea, create a gamut of t statistics to get the
empirical distribution of the null.
#{| stat || obs |
p value
# samples
Permutation Test Contd
• We should ONLY use permutation tests when the number
of replicates are >10, else the number of possible
resamples is quite small.
• Example when n1=4, n2=4. The possible permutation is
• 8!/4!4!=70 possibilities, which is too small to estimate
empirical p-values.
• Also, we need a firm assumption that the ONLY difference
between the two samples is due to location and they have
the same shapes.
Wilcoxon Rank Sum Test
• The non-parametric alternative to the two-sample t.
• Doesn’t assume underlying normality
• Pool the data and rank all the observations in the pooled
data.
• Then sum the ranks of ONE of the samples.
• Idea is, if there is a location difference among the samples,
the sum of ranks for one sample will be either bigger or
smaller than the other. Wilcoxon Rank Sum tables are
generally available.
• Not very good for VERY few replicates. At least 4,
preferably 6.
Large Sample Approx.
• Here n1 is size of the sample we summed
up.
W n1 (n2 n1 1) / 2
n2n1 (n2 n1 1) /12
Likelihood Ratio Test
• The idea of Likelihood Ratio Test is simple and intuitive.
• One basically looks at the ratio of the likelihood that the
gene is differentially expressed to that of the likelihood it
isn’t.
• The method was used by Idekar et al (2000).
• Model:
xg1k = g1 + g1eg1k + dg1k
•
xg2k = g2 + g2eg2k + dg2k
• Allows both additive and multiplicative error.
Assumption, egik, eg2k = BVN( 0, 0, s2eg1, s2eg2, re)
dg1k, dg2k = BVN( 0, 0, s2dg1, s2dg2, rd)
• Allows correlations.
The LR Statistic
• The parameters
• b = (s2eg1, s2eg1, re,s2dg1, s2dg1, rd) and =(g1,g2) are
estimated via ML techniques optimizing:
n
K
P( x
g 1k
,xg1k | b , )
g 1 k 1
The LR Statistic is
g 2 ln(
max g Lg ( b , g1 g 2 g )
max g1, g 2 Lg ( b , g1 , g 2 )
12
The Issues of MULTIPLE Testing
• In Microarray setting there are often thousands of tests to
be considered simultaneously.
• Obviously we have a pretty high chance of having false
positives when you do thousands of tests at the same time.
• In these kinds of situations, the question of which error to
control becomes an issue.
• We would like to control both Type I and Type II errors,
but that’s not possible
– If you make Type I error smaller, Type II goes up and vice versa.
The general practice is to fix Type I error at a low level (.05, .01, .001)
and minimize the Type II error for that level.
Numbers of Correct and Incorrect Decisions
Truly
Inactive
Truly
Active
Total
Declared
Inactive
U
Declared
Active
V =FP
Total
T=FN
S=TP
n-n0
n-R
R
n
n0
What Error Rates do we need to control?
• Most traditional methods focus on controlling the
expected fraction of false positives out of the total number
of TRUE null hypotheses.
• We want the expected fraction of false positives to be
small. We want to control
• FPR = E ( FP / n0)
• However since we do not know n0 we really cannot control
this rate. So we look at several measures that are related to
this rate.
Per-comparison Error Rate (PCER):
• PCER = E(V)/n
• Controlling the expected number of False positives as a
fraction of the total number of hypothesis tested.
• Here a level control achieved by performing each test in
the family at a level alpha
• error arising from multiplicity ignored
• false claims of significance are made at a higher rate than
Per-family Error Rate (PFER):
• PFER = E(V)
• The other end of the spectrum: controls the expected
number of False positive per each hypothesis tested.
• This is the expected number of False Positives in a study.
• procedures that control this error rate are very strict
• rarely used in microarray literature
Family-wise Error Rate (FWER):
• FWER = P(V ≥ 1)
Controls the probability of having a single Type I error in the
study. Most commonly used, common examples are:
Bonferroni, Tukey, Dunnett
• procedures based on this error rate are very conservative
• generalized version of this error rate: k-FWER
False Discovery Rate (FDR):
• FDR = E(V/R | R ≥ 1)
• Controls the expected number of False positives OUT OF
THE ONES DECLARED POSITIVE.
• FDR controlling procedures are liberal
• widely used in microarray literature
The Methods Compared
• The hierarchy among techniques
• PCER ≥ FDR ≥ FWER ≥ PFER
• PCER is the least conservative and PFER is the most
conservative.
• These days people use either FWER or FDR
• These are all SINGLE STEP methods. However, Stepwise methods are getting more popular.
Stepwise methods
• Essentially a more adaptive form of testing.
• We can have step-down methods or step-up methods.
• Step-down methods: rank observed p-values from the
smallest to the highest,
– p(1)≤ … ≤p(m)
– Start at the smallest p-value, p(1).
• Step-Up Method
– Rank as before
– start with the largest p-vlaue
Step-Down: Bonferroni Holm Method
• rank observed p-values from the smallest to the highest,
• p(1)≤ … ≤p(m) and define the corresponding H1….Hm
Start with the smallest one, reject H1 if p(1) < a/m and
continue to Step 2 using p(2) and so on…
Else Fail to reject all H1….Hm.
• And reject H(j) if
• p(j)<a/(m - j + 1)
• This is more powerful than single-step procedures
Step-up: Hochberg and Hommel
• rank observed p-values from the smallest to the highest,
• p(1)≤ … ≤p(m) and define the corresponding H1….Hm
Start with the LARGEST one, reject Hm if p(m) < a and all the
corresponding H1….Hm
Otherwise Fail to reject and continue to Step 2 using p(m-1) and so on…
• And reject H(j) if
• this procedure rejects H(1),...,H(k)
• if k =Max{j| p(j) ≤ ja/m} exists.
• This is more powerful than single-step procedures and the Step-Down
procedure and controls FDR
Error Rates: used in microarrays
• FWER Bonferroni: Classify all genes as active if their p-value is less
than a/n
• Step-up FWER(Hochberg): Let k be the largest g =1,…n, for which
p(g) <= a/ (n-g+1). Then reject all H(g) for g=1…k where H(g) is the
null associated with the gth smallest p-value.
• Step-down FWER(Holmes): Let k be the smallest g =1,…n, for
which p(g) >= a/ (n-g+1). Then fail to reject all H(g) for g=1…k where
H(g) is the null associated with the gth largest p-value
• FDR(Benjamini and Hochberg): Let k be the largest g =1,…n, for
which p(g) <= ag/np0). Then reject all H(g) for g=1…k where H(g) is the
null associated with the gth smallest p-value. (here p0 is the true
proportion of inactive genes which is unknown, and generally replaced
by 1 a conservative value)