022107
advertisement

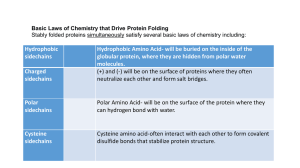
Couple of lab things 1. In calculating rate (mmol/min/mg) from absorbance/time slope, take the absolute value (rates always (usually?) positive) 2. For write-up: in your table of rates, please include the raw data (Dabs/time) as well as the calculated rate +1 +2 0 + - + + +1.5 + -1 0 + +0.5 - 0 0 -0.5 0 pI between pKR and pK2 Free amino acid vs. polymers Pentapeptide (five a carbons!) What’s the charge at pH=7? Different polypeptides have different charge characteristics 1. Amino terminus (if ‘free’) 2. Carboxyl terminus 3. Charged side chains Calculated pI ~7.0 http://www.embl-heidelberg.de/cgi/pi-wrapper.pl • Free amino acids vs. polymerized – Side chains may have different pKas • pKa affected by charges on amino/carboxyl groups • pKa may be affected by interactions with other side chains in the larger molecule Nucleophilic B: strengthens the acidic character of the adjacent serine Look at protein structure at a low resolution • Primary (1°) structure – Sequence of amino acids • Total number: anywhere from two to tens of thousands – Few (2 to tens) amino acids: oligopeptides » Typically hormones, etc. – Hundreds or more » “Proteins” » Enzymes, structural proteins, etc. Look at protein structure at a low resolution • Non-amino acid chemical groups can enhance protein function – “Prosthetic groups” “Enzyme co-factor” – Associated or covalently-bound – eg. Metals • Iron, Calcium, Zinc, Magnesium, etc. • Structural components • Good nucleophiles: enzymatic ‘activation’ of water, for example • Redox chemistry: accept/donate electrons Look at protein structure at a low resolution • Prosthetic groups – Lipids (lipoproteins) or sugars (glycoproteins) • Enhance protein stability • Alter interactions with other biological molecules http://www.liv.ac.uk/physiology/ncs/conform.html Look at protein structure at a low resolution • Primary (1°) structure • Secondary (2°) structure – Arrangement of local stretches of amino acids – Stabilized predominantly by hydrogen bonds between backbone N-H and O=C Common elements of 2° structure a helix b strand Look at protein structure at a low resolution • Primary (1°) structure • Secondary (2°) structure • Tertiary (3°) structure – Three-dimensional fold of a polypeptide – How do 2° structure elements interact? • Non-covalent interactions – Hydrophobic interactions – H-bonds – van der Waals • Covalent bonds: disulfide a-helices interact to give the overall 3D fold of a polypeptide Look at protein structure at a low resolution • • • • Primary (1°) structure Secondary (2°) structure Tertiary (3°) structure Quaternary (4°) structure – Interaction between subunits of a multisubunit protein • Non-covalent interactions • Disulfide (covalent) bonds SUBUNITS (INDIVIDUAL POLYPEPTIDES) PROTEIN Different proteins have different chemical characteristics • Caused by 1°, 2°, 3°, 4° structures • Define their biological roles • Can be exploited to separate proteins from each other – Purify a single protein of interest Different proteins have different chemical characteristics 1. Charge • Isoelectric point • • At pH > pI, net charge ? 0 At pH < pI, net charge ? 0 2. Size • • Sum of masses of all amino acids (minus 18) Effective size can be influenced by 3°, 4° structure Effective sizes can be influenced by protein shape Globular Filamentous Different proteins have different chemical characteristics 3. Ligand-binding/Affinity for other molecules 4. Exposed hydrophobic patches Exploitation of chemical characteristics • Purification – Chromatography • Separation/analysis – Electrophoresis