H 2 - Milwaukie High School
advertisement

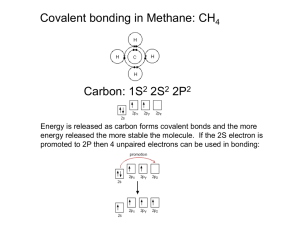
Molecular Geometry and Bonding Theories The properties of a molecule depend on its shape and and the nature of its bonds. In this unit, we will discuss three models. (1) a model for the geometry of molecules -- valence-shell electron-pair repulsion (VSEPR) theory (2) a model about WHY molecules form bonds and WHY they have the shape they do -- valence-bond theory (3) a model of chemical bonding that deals with the electronic structure of molecules -- molecular orbital (MO) theory bond angles: the angles made by the lines joining the nuclei of a molecule’s atoms carbon dioxide CO2 180o methane CH4 109.5o formaldehyde CH2O 120o VSEPR electron domain: a region in which at least two electrons are found -- they repel each other because… they are all (–) bonding domain: 2-to-6 e– that are shared by two atoms; they form a… covalent bond nonbonding domain: 2 e– that are located on a single atom; also called a… lone pair For ammonia, there are NH3 .. three bonding domains and H– N –H one nonbonding domain. .. H N H Domains arrange themselves so H H as to minimize their repulsions. .. The electron-domain geometry is one of five basic arrangements of domains. N H H H -- it depends only on the total # of e– domains, NOT the kind of each domain The molecular geometry describes the orientation of the atoms in space. -- it depends on how many of each kind of e– domain .. Total # of Electron-Domain Domains Geometry 2 3 4 5 6 linear Possible Molecular Geometries linear (CO2) trigonal planar trigonal planar (BF3), bent (NO2) tetrahedral tetrahedral (CH4), trigonal pyramidal (NH3), bent (H2O) trigonal bipyramidal trig. bipyramidal (PCl5), linear (XeF2) seesaw (SF4), T-shaped (ClF3) octahedral octahedral (SF6), sq. pyr. (BrF5), square planar (XeF4) “atoms – axial” To find the electron-domain geometry (EDG) and/or molecular geometry (MG), draw the Lewis structure. Multiple bonds count as a single domain. Predict the EDG and MG of each of the following. SeCl2 20 e– .. .. .. .. .. Cl–Se–Cl .. .. .. .. .. 18 e– ] – .. O3 [ .. .. 26 e– .. SnCl3 – .. .. .. Cl– .. Sn–Cl .. Cl .. .. O– .. EDG: tetrahedral MG: trig. pyramidal EDG: trig. planar MG: bent EDG: tetrahedral MG: bent IF5 EDG: trig. planar MG: trig. planar .. .. .. .. .. .. .. .. 34 e– 2– ] F .. .. SF4 [ .. 24 e– (res.) CO32– .. O– C= O .. O .. .. .. F .. ..F– S –F .. ..F .. EDG: octahedral MG: sq. pyramidal I 42 e– EDG: trig. bipyr. MG: seesaw 28 e– .. .. .. ClF3 .. .. .. ..F .. .. .. ..F– Cl –F .. ..F EDG: trig. bipyr. MG: T-shaped .. .. .. [ – ] .. Cl .. .. .. 36 e– .. ICl4– Cl .. .. Cl– .. I –Cl .. EDG: octahedral MG: sq. planar For molecules with more than one central atom, simply apply the VSEPR model to each part. Predict the EDG and MG around the three interior atoms of ethanoic (acetic) acid. CH3COOH PORTION EDG MG H O .. H–C–C–O–H .. H –CH3 tetra. tetra. C=O trig. plan. trig. plan. .. tetra. bent –OH .. Nonbonding domains are attracted to only one nucleus; therefore, they are more spread out than are bonding domains. The effect is that nonbonding domains (i.e., “lone pairs”) compress bond angles. Domains for multiple bonds have a similar effect. e.g., the ideal bond angle for the tetrahedral EDG is 109.5o H H H2O N O .. C H 109.5o H H H H COCl2 124.3o H O=C 124.3o 107.0o 104.5o .. Cl .. ..111.4 Cl .. .. NH3 o .. CH4 H EDG = trig. plan. ideal = 120o Polarity of Molecules A molecule’s polarity is determined by its overall dipole, which is the vector sum of the dipoles of each of the molecule’s bonds. Consider CO2 v. H2S... bond dipoles O=C=O bond dipoles S H H overall dipole = zero overall dipole = (NONPOLAR) (POLAR) Classify as polar or nonpolar: BCl3 .. .. .. Cl B .. .. 26 e– .. PCl3 .. .. .. Cl– .. P–Cl .. Cl .. .. 24 e– Boron can be extracted by the electrolysis of molten boron trichloride. Boron is an essential nutrient for plants, and is also a primary component of control rods in nuclear reactors. polar nonpolar Valence-Bond Theory: merges Lewis structures w/the idea of atomic orbitals (2s, 3p, etc.) Lewis theory says… covalent bonding occurs when atoms share electrons V-B theory says… covalent bonding occurs when valence orbitals of adjacent atoms overlap; then, two e–s of opposite spin (one from each atom) combine to form a bond V-B theory is like Velcro: “No overlap, no bond.” Consider H2, Cl2, and HCl... (unpaired e– is in 1s orb.) H Cl [Ne] H2 (unpaired e– is in 3p orb.) Cl2 HCl = orbital overlap region (responsible for bond) There is always an optimum distance between two bonded nuclei. At this optimum distance, attractive (+/–) and repulsive (+/+ and –/–) forces balance. Energy 0 H2 molecule optimum dist. (min. PE) H–H distance Hybridized Orbitals V-B theory can’t explain some observations about molecular compounds without the concept of hybridized orbitals. i.e., covalent (NM/NM) compounds :F Be F: : : BF3 : : BeF2 : : In… (LS) …the central atom …but is ACTUALLY SHOULD be… hybridized to be… 2s 2p sp 2s 2p sp2 2p 2 hyb. sp orbs. 2 unhyb. 2p orbs. 3 hyb. sp2 orbs. 1 unhyb. 2p orb. 2p B F: H2O H O: H 2s 2p 4 hyb. sp3 orbs. sp3 : CH4 H H C H H …the result then being… : : PF5 2s P F: XeF4 :F: :F Xe F: :F: : : :F Xe F: : : : XeF2 3s 5s 2p 3p 5p 4 hyb. sp3 orbs. sp3 3d 5d sp3d sp3d 3d 5 hyb. sp3d orbs. 4 unhyb. 3d orbs. 5d 5 hyb. sp3d orbs. 4 unhyb. 5d orbs. : : : : : : 5s 5p 5d sp3d2 5d 6 hyb. sp3d2 orbs. 3 unhyb. 5d orbs. KEY: EDG hybridization of central atom linear sp trig. planar sp2 tetrahedral sp3 trig. bipyr. sp3d octahedral sp3d2 sp hybridization sp hybridization occurs when one s and one of the three p orbitals hybridize. It results in the creation of two sp hybrid orbitals (orange). The other two p orbitals (purple) are unhybridized; they retain the “figure-8” or “dumbbell” shape. Multiple Bonds s (sigma) bonds are bonds in which the e– density is .. along the internuclear axis. N -- These are the single bonds we have H H H considered up to this point. -- e.g., s-s, s-p, or p-p overlap, The bonds and also p-sp hybrid overlap in ammonia are s bonds. sigma (s) = single Multiple bonds result from the overlap of two p orbitals (one from each atom) that are oriented perpendicularly to the internuclear axis. These are p (pi) bonds. pi, multiple, unhybridized p bonds are generally weaker than s bonds because p bonds have less overlap. For ethene (C2H4)… H H H C=C H H For each carbon atom, there are 3 sp2 hybridized orbitals; these form s H bonds (- - - -) with C or H. Where unhybridized orbitals overlap, a p bond ( ) is formed. H C C H Single bonds are s bonds. e.g., C2H6 Double bonds consist of one s and one p. e.g., C2H4 Triple bonds consist of one s and two p. e.g., C2H2 Experiments indicate that all of C2H4’s atoms lie in the same plane. This suggests that p bonds introduce rigidity (i.e., a reluctance to rotate) into molecules. -- p bonding does NOT occur with sp3 hybridization, only sp and sp2 -- p bonding is more prevalent with small molecules (e.g., C, N, O) (Big atoms don’t allow enough p-orb. overlap for p bonds to form.) Delocalized p Bonding Localized p bonds are between… two atoms only. -- These are common for molecules with… resonance structures. .. .. Delocalized p bonds are “smeared out” and shared among… > two atoms. [ .. O– N=O .. O .. .. -- e.g., C2H2, C2H4, N2 ] – The nitrate ion (NO3–) has delocalized electrons. -- The electrons involved in these bonds are delocalized electrons. H Consider benzene, C6H6. -- Each carbon atom is sp2 hybridized. ___ H C H C C C -- This leaves... a 2p orbital on each C C C that is oriented H H to plane of m’cule. -- 6 e– shared equally by 6 C atoms H Which of the following exhibit delocalized bonding? = – – = – – H O H H–C–C–C–H H H propanone .. SO2 18 e– sulfur dioxide .. O–S=O .. “YEP.” (res.) .. .. .. [ .. O .. O– S –O .. .. O .. ] .. .. .. sulfate ion 32 e– .. SO4 2– “NOPE.” .. O H3C–C–CH3 24 e– 2– “NOPE.” Molecular Orbitals -- The VSEPR and valence-bond theories don’t explain the excited states of molecules, which come into play when molecules absorb and emit light. -- This is one thing that the molecular orbital (MO) theory attempts to explain. Molecules respond to the many wavelengths of light. The wavelengths that are absorbed and then re-emitted determine an object’s color, while the wavelengths that are NOT re-emitted raise the temperature of the object. molecular orbitals: wave functions that describe the locations of electrons in molecules -- these are analogous to atomic orbitals in atoms (e.g., 2s, 2p, 3s, 3d, etc.), but MOs are possible locations of electrons in molecules (not atoms) -- MOs, like atomic orbitals, can hold a maximum of two e– with opposite spins -- but MOs are for entire molecules MO theory is more powerful than valence-bond theory; its main drawback is that it isn’t easy to visualize. Hydrogen (H2) The overlap of two atomic orbitals produces two MOs. (antibonding MO) s*1s 1s + 1s H atomic orbitals s1s (bonding MO) H2 molecular orbitals -- The lower-energy bonding molecular orbital concentrates e– density between nuclei. -- For the higher-energy antibonding molecular orbital, the e– density is concentrated outside the nuclei. -- Both of these are s molecular orbitals. Energy-level diagram (molecular orbital diagram) s*1s 1s 1s H atom s1s H atom H2 m’cule (antibonding MO) s*1s 1s + 1s H atomic orbitals s1s (bonding MO) H2 molecular orbitals Consider the energy-level diagram for the hypothetical He2 molecule… s*1s 1s 1s He atom s1s He atom He2 m’cule 2 bonding e–, 2 antibonding e– No energy benefit to bonding. He2 molecule won’t form. bond order = ½ (# of bonding e– – # of antibonding e–) -- the higher the bond order, the greater the bond stability 0 = no bond 1 = single bond 2 = double bond 3 = triple bond 007 7 = James Bond -- MO theory allows for fractional bond orders as well. -- a bond order of... What is the bond order of H2+? 1 e– total 1 bonding e–, zero antibonding e– BO = ½ (1–0) = ½ Second-Row Diatomic Molecules 3 4 Li Be 6.941 9.012 B 5 10.811 C 6 12.011 7 8 N O 14.007 15.999 F 9 18.998 10 Ne 20.180 1. # of MOs = # of combined atomic orbitals 2. Atomic orbitals combine most effectively with other atomic orbitals of similar energy. 3. As atomic orbital overlap increases, bonding MO is lowered in energy, and the antibonding MO is raised in energy. 4. Both the Pauli exclusion principle and Hund’s rule apply to MOs. Use MO theory to predict whether Li2 and/or Be2 could possibly form. s*2s 2s 2s s2s s*1s 1s 1s Li BO = ½ (4–2) = 1 s1s Li Li2 “YEP.” s*2s 2s 2s s2s Bonding and antibonding e– cancel each other out in core energy levels, so any bonding is due to e– in bonding orbitals of outermost shell. Be Be BO = ½ (4–4) = 0 Be2 “NOPE.” Molecular Orbitals from 2p Atomic Orbitals The 2pz orbitals overlap in head-to-head fashion, so these bonds are... s bonds. -- the corresponding MOs are: s2p and s*2p y x z The other 2p orbitals (i.e., 2px and 2py) overlap in sideways fashion, so the bonds are... p bonds. -- the corresponding MOs are: p2p (two of these) and p*2p (two of these) Rule 3 above suggests that, from low energy to high, the 2p MOs SHOULD follow the order: LOW HIGH s2p < p2p < p*2p < s*2p ENERGY ENERGY General energy-level diagrams for MOs of second-row homonuclear diatomic molecules... don’t fit on this slide. (And so, they’re on the next one… …if that’s all right with you.) (And if not, you can… …quit and go make pancakes.) For B2, C2, and N2... s*2p “Mr. B” (or Mr. C) (or Mr. N) same for both p*2p s2p p2p For O2, F2, and “Ne2”... s*2p p*2p p2p s2p s*2s s*2s s2s s2s (1s MOs are down here) Here, the interaction is weak. Here, the interaction between the 2s of one atom and the 2p The energy distribution is as expected. of the other is strong. The orbital energy distribution is altered. paramagnetism: describes the attraction of molecules with unpaired e– to a magnetic field diamagnetism: describes substances with no unpaired e– (“di-” = two; diamagnetic ~ = “dielectron”) -- such substances are VERY weakly (almost unnoticeably) repelled by a magnetic field Use the energy diagrams above to tell if diatomic species are paramagnetic or diamagnetic. paramagnetism of liquid oxygen Paramagnetic or diamagnetic? B2 (10) P C2 (12) D N2 (14) D s*2p p*2p s2p p2p s*2p p*2p p2p s2p s*2s s*2s s2s s2s O2 (16) P F2 (18) D s*1s s*1s O2+ (15) P s1s s1s O22– (18) D C22– (14) D