A cell free phosphorylation method to assess the utility of new
advertisement

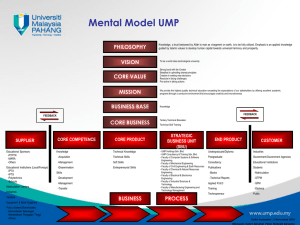
A cell free phosphorylation method to assess the utility of new nucleotides as Nucleotide Reverse Transcriptase Inhibitors Phiyani J. Lebea, Moira L. Bode, Kgama Mathiba and Dean Brady CSIR Biosciences, Ardeer Rd., Modderfontein, 1645, South Africa Background and Objectives The acyclic nucleoside phosphonates such as tenofovir have proved to be effective against a wide variety of DNA virus and retrovirus infections1. Generally, phosphonates need to be converted into a prodrug to improve their bioavailability. Secondly, they have to be recognised by the host cell kinases for subsequent phosphorylation into triphosphates. Only after both of these two steps will they be effective as HIV-1 reverse transcriptase inhibitors. Therefore, the development of assays that can separate bioavailability from phosphorylation and enzyme inhibition are critical for high throughput screening of potential nucleotide reverse Transcriptase Inhibitors (NtRTIs). The objective of this study was to evaluate the cell-free phosphorylation of tenofovir and UMP, as test cases, using mammalian cell extracted nucleotide kinases. Further studies would then concentrate on cell-free phosphorylation of new compounds prepared as possible reverse transcriptase inhibitors to assess their capacity for phosphorylation as well as linking this directly to testing HIV-1 reverse transcriptase enzyme inhibition potential in a single assay. Figure 3 β-NADH depletion as a function of nucleotide monophosphate phosphorylation to its triphosphate product Methods Control (No enzym e) vs Assay control (UMP) Intact Caco2 and HeLa cells were fractionated into cytosolic and membrane proteins and the total compartmentalized native proteins were quantified. Substrate (monophosphate and phosphonate) phosphorylation was initiated by addition of the isolated total protein mixture with pyruvate kinase as the secondary phosphorylation catalyst. Lactate dehydrogenase was used in the reaction to catalyse conversion of the resultant pyruvate to lactate with concomitant oxidation of β-NADH cofactor2. The reaction scheme is shown in figure 1. The amounts of mono-, di- and triphosphate present in the reaction mixture were quantified using HPLC. 1.22 1.2 Optical density 1.18 Figure 1 Reaction scheme of the phosphorylation of phosphonates and natural nucleoside monophosphates y = -0.0044x + 1.206 R2 = 0.9676 1.16 1.14 1.12 y = -0.0231x + 1.1935 R2 = 0.9986 1.1 1.08 ATP + NMP NMP kinase (cellular protein mixture) ADP + NDP (1) 1.06 0 1 2 3 4 5 6 Time (minutes) Control ADP + NDP + 2PEP Pyruvate kinase ATP + NTP + 2Pyruvate UMP Linear (Control) Linear (UMP) (2) Figure 4 2Pyruvate + (2NADH + H+) 2Lactate + 2NAD+ LDH β-NADH depletion as a function of nucleotide monophosphate (UMP) and phosphonate (tenofovir) phosphorylation to their respective triphosphate products (3) Beta-NADH depletion as a function of nucleoside phosphorylation 1.2 1.18 Figure 2 1.16 Optical density Wavelength scan (beta-NADH) 5 4.5 y = -0.0146x + 1.1925 R2 = 0.9844 1.14 1.12 1.1 y = -0.0191x + 1.1754 R2 = 0.9919 4 Optical density 1.08 3.5 1.06 3 0 1 2 3 2.5 4 5 6 Time (minutes) Tenofovir 2 UMP Linear (Tenofovir) Linear (UMP) 1.5 1 Figure 5 0.5 0 0 50 100 150 200 250 300 350 400 450 HPLC quantification chromatograms of the phosphorylation products of tenofovir and UMP (insert). The left panel indicates diphosphate formation from tenofovir and UMP. The right panel shows triphosphate product yields of tenofovir and UMP. 7.680 - ATP mAU 160 120 140 5.109 Tenofovir diphosphate 7.682 ATP 100 100 40 References 40 20 0 0 7.696 - ATP 60 7.725 ATP 80 20 20 The first experiment was conducted using only the extracted total cellular proteins to confirm the phosphorylation of the primary substrates (UMP and tenofovir) to their diphosphates (reaction 1, figure1). Figure 5 shows the presence of tenofovir diphosphate and uridine diphosphate (dotted lines) in significant quantities when compared to the blank sample (solid lines). In the second experiment pyruvate kinase (reactions 1 & 2, figure 1) was added together with the cellular total protein mixture to convert the diphosphate into the triphosphate form. Although UMP is converted to UTP, tenofovir is mainly converted to its diphosphate and not to the triphosphate as shown in figure 5. The lack of triphosphorylation of tenofovir is due to competition with ADP as a substrate which is preferentially phosphorylated over the artificial nucleoside analogue. 100 5.112 - ADP 40 120 7.191 - UTP 5.071 - ADP 20 60 7.701 ATP 80 140 4.831 - UDP 100 mAU 3.391 - UMP 60 5.108 - ADP 40 4.660 - Tenofovir diphosphate 3.392 - UMP 60 mAU 120 80 7.683 - ATP 80 120 5.067 - ADP The optimal wavelength with which to monitor β-NADH oxidation was determined to be 340 nm as shown in figure 2. The β-NADH was monitored at 340 nm throughout the initial 5 minutes of the reaction to compute the reaction turnover and thus calculate the activity of the nucleotide monophosphate kinase. The phosphorylation of both the natural substrate (UMP) and tenofovir using the extracted total cytoplasmic kinases as well as utilising membrane bound total proteins was shown to be possible since there was considerable oxidation of β-NADH compared to the control sample (figure 3 and 4). The cytoplasmic reaction rate of substrate phosphorylation was found to be 2.7 x 10-2 for the natural substrate uridine monophosphate (UMP) while that of tenofovir was 1.7 x 10-2 units/ml of enzyme. Furthermore the cytosolic conversion rate of the natural substrate (UMP) was 20% faster than that of the same amount of protein extract from the membrane bound total protein. 7.709 ATP 3.362 - Tenofovir mAU 7.676 - ATP Results and Discussion 3.363 - Tenofovir Wavelength (nm) 0 2 4 6 8 10 12 14 min 0 0 2 4 6 8 10 12 14 min 0 0 2 4 6 8 10 12 14 min 0 2 4 6 8 10 12 14 Conclusions Cell free phosphorylation of mono- and diphosphate nucleotides as well as phosphonates has been shown to be possible, providing a method for assessing the suitability of newly designed compounds as possible NtRTIs in a cell free system. Relative rates of phosphorylation between the cytosol and mitochondria can be determined and used as a means of evaluating the mitochondrial capacity to process the inhibitor compounds. Furthermore, even before the newly synthesized phosphonates are evaluated as possible inhibitors of the HIV-1 reverse transcriptase, their potential phosphorylation in the cells of choice can be tested... 1. De Clercq, E. (1997) Acyclic nucleoside phosphonates in the chemotherapy of DNA virus and retrovirus infection. Intervirology 40 (5 – 6): 295 – 303 2. Blondin, C., Serina, L., Wiesmuller, L., Gilles, A.-M. and Barzu, O. (1994). Improved spectrometric assay of nucleoside monophosphate kinase activity using the pyruvate kinase /lactate dehydrogenase coupling system. Anal Biochem. 220:219-221 min